- Home
- Products
- Services
- Resources
- About Us
- News
- Contact

loading
CBPE0390
CBPE0390
| Availability: | |
|---|---|
Description
DNA methylation is an epigenetic modification, a form of chemical modification of DNA, which refers to the carbon 5 of cytosine in the genomic CpG dinucleotide under the action of DNA methyltransferase. covalently bonded to a methyl group. It can change genetic expression without changing the DNA sequence.
General information
Cat No. | Name | Average Methylation Level | Description |
CBPE0390-1 | Human Non-Methylation Reference Standard | Average Methy=0% | This product is obtained by whole genome amplification (WGA) and can be used as a negative control for DNA methylation analysis. The test method is verified by RRBS, ddPCR and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-2 | Human Low-Methylation Reference Standard | Average Methy <30% | The product is obtained by knocking out DNMT3A and DNMT3B proteins at the cellular level and extracting gDNA. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-3 | Human Medium-Methylation Reference Standard | 30% | This product is obtained by overexpressing DNMT3A and DNMT3B proteins in cells and extracting gDNA. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-4 | Human High-Methylation Reference Standard | Average Methy>70% | This product is a natural sample. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-5 | Human Hyper-Methylation Reference Standard | Average Methy≈100% | The whole genome methylated DNA of this product is obtained by modifying human genomic DNA with methyltransferase (M.SssI), which can be used as a positive control for DNA methylation analysis. The test method is verified by RRBS, ddPCR and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
Remark | 1. MspI can digest both unmethylated and methylated DNA, but is insensitive to methylation. 2. HpaII is sensitive to CpG methylation, but cannot digest methylated DNA, only unmethylated DNA. 3. This product uses the same cell line | ||
Storage Conditions | 2~8℃ | ||
Expiry | 36 months from the date of manufacture | ||
Representative Data
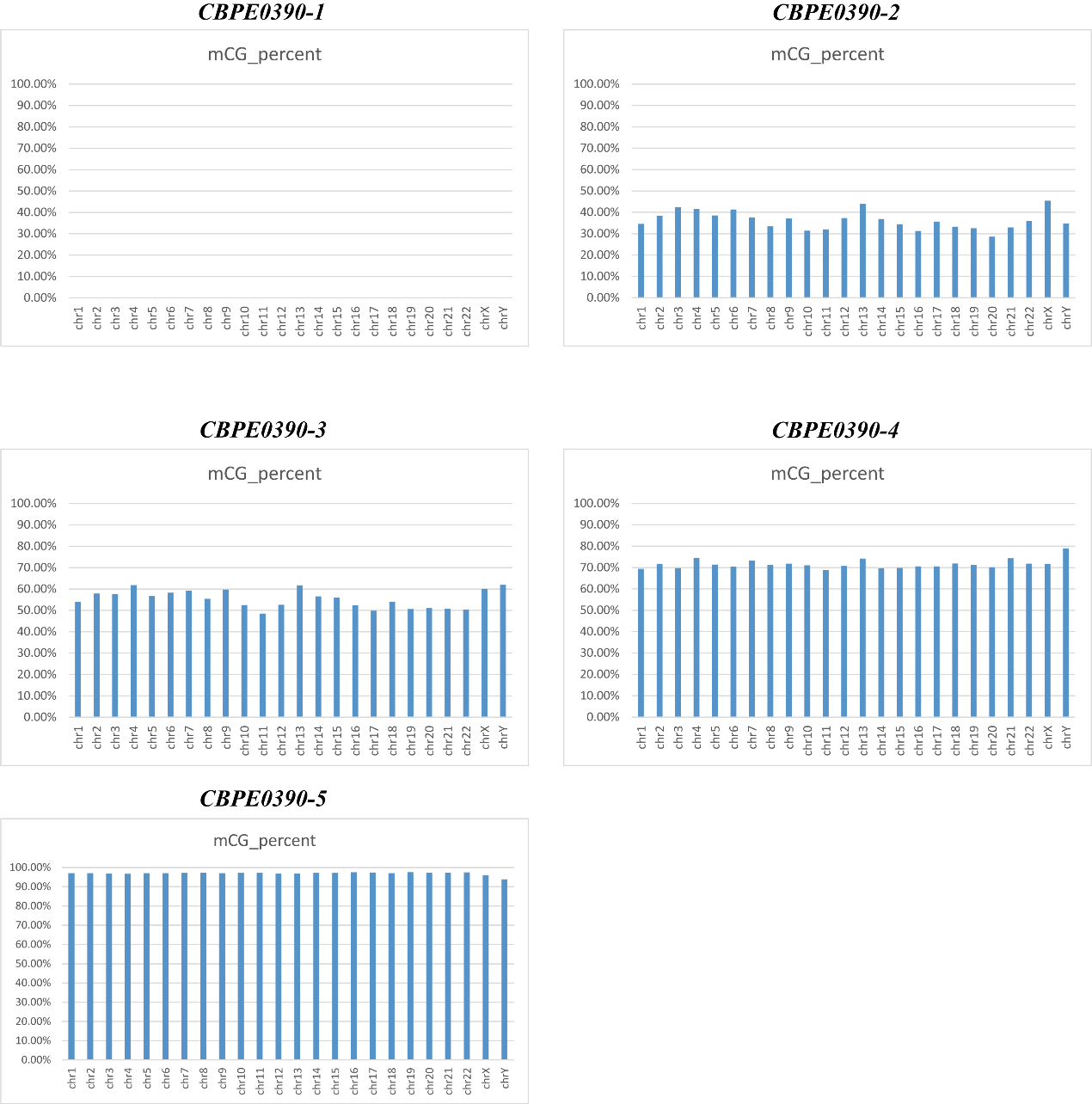
Figure 1. DNA methylation analysis by Reduced representation bisulfite sequencing (RRBS) on different samples.
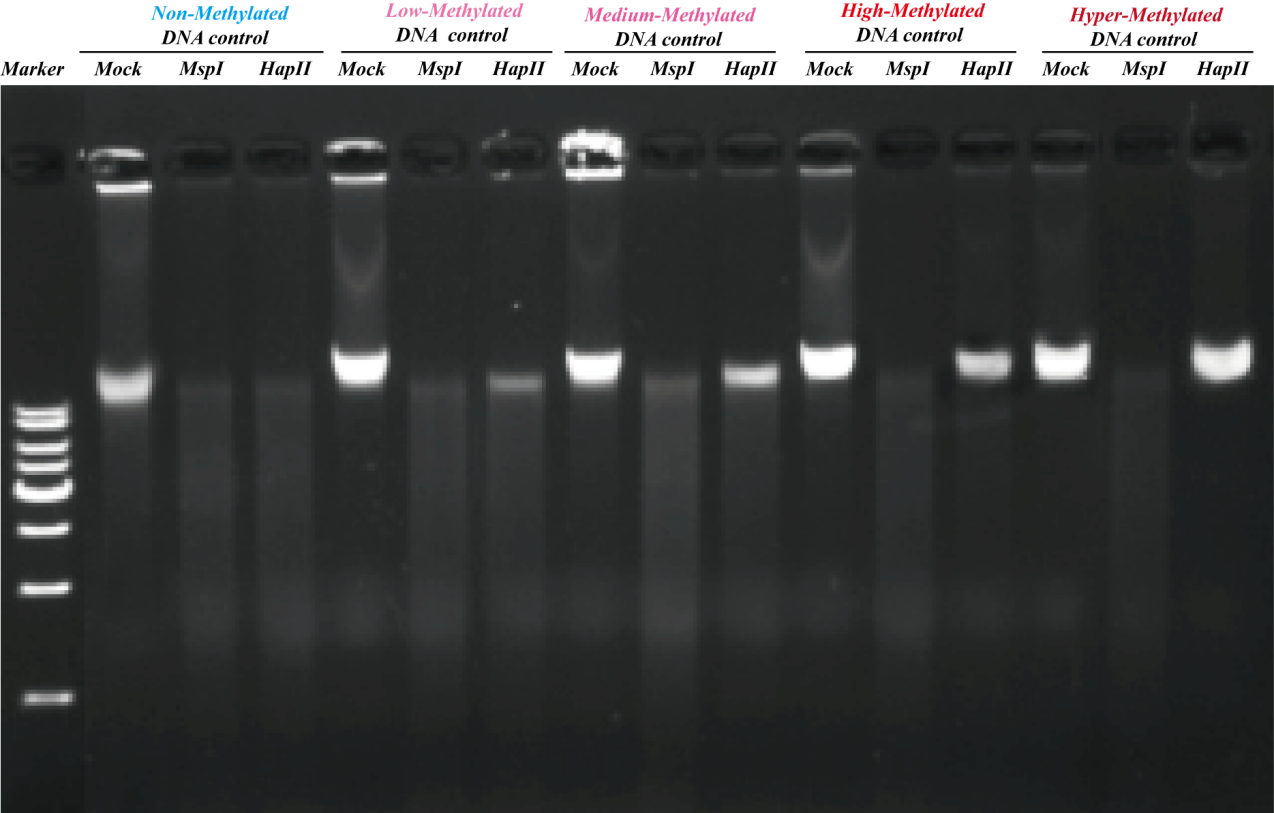
Figure 2. Different samples digested by MspI or HapII.
Description
DNA methylation is an epigenetic modification, a form of chemical modification of DNA, which refers to the carbon 5 of cytosine in the genomic CpG dinucleotide under the action of DNA methyltransferase. covalently bonded to a methyl group. It can change genetic expression without changing the DNA sequence.
General information
Cat No. | Name | Average Methylation Level | Description |
CBPE0390-1 | Human Non-Methylation Reference Standard | Average Methy=0% | This product is obtained by whole genome amplification (WGA) and can be used as a negative control for DNA methylation analysis. The test method is verified by RRBS, ddPCR and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-2 | Human Low-Methylation Reference Standard | Average Methy <30% | The product is obtained by knocking out DNMT3A and DNMT3B proteins at the cellular level and extracting gDNA. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-3 | Human Medium-Methylation Reference Standard | 30% | This product is obtained by overexpressing DNMT3A and DNMT3B proteins in cells and extracting gDNA. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-4 | Human High-Methylation Reference Standard | Average Methy>70% | This product is a natural sample. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-5 | Human Hyper-Methylation Reference Standard | Average Methy≈100% | The whole genome methylated DNA of this product is obtained by modifying human genomic DNA with methyltransferase (M.SssI), which can be used as a positive control for DNA methylation analysis. The test method is verified by RRBS, ddPCR and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
Remark | 1. MspI can digest both unmethylated and methylated DNA, but is insensitive to methylation. 2. HpaII is sensitive to CpG methylation, but cannot digest methylated DNA, only unmethylated DNA. 3. This product uses the same cell line | ||
Storage Conditions | 2~8℃ | ||
Expiry | 36 months from the date of manufacture | ||
Representative Data

Figure 1. DNA methylation analysis by Reduced representation bisulfite sequencing (RRBS) on different samples.

Figure 2. Different samples digested by MspI or HapII.
General information
Cat No. | Name | Average Methylation Level | Description |
CBPE0390-1 | Human Non-Methylation Reference Standard | Average Methy=0% | This product is obtained by whole genome amplification (WGA) and can be used as a negative control for DNA methylation analysis. The test method is verified by RRBS, ddPCR and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-2 | Human Low-Methylation Reference Standard | Average Methy <30% | The product is obtained by knocking out DNMT3A and DNMT3B proteins at the cellular level and extracting gDNA. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-3 | Human Medium-Methylation Reference Standard | 30% | This product is obtained by overexpressing DNMT3A and DNMT3B proteins in cells and extracting gDNA. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-4 | Human High-Methylation Reference Standard | Average Methy>70% | This product is a natural sample. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-5 | Human Hyper-Methylation Reference Standard | Average Methy≈100% | The whole genome methylated DNA of this product is obtained by modifying human genomic DNA with methyltransferase (M.SssI), which can be used as a positive control for DNA methylation analysis. The test method is verified by RRBS, ddPCR and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
Remark | 1. MspI can digest both unmethylated and methylated DNA, but is insensitive to methylation. 2. HpaII is sensitive to CpG methylation, but cannot digest methylated DNA, only unmethylated DNA. 3. This product uses the same cell line | ||
Storage Conditions | 2~8℃ | ||
Expiry | 36 months from the date of manufacture | ||
General information
Cat No. | Name | Average Methylation Level | Description |
CBPE0390-1 | Human Non-Methylation Reference Standard | Average Methy=0% | This product is obtained by whole genome amplification (WGA) and can be used as a negative control for DNA methylation analysis. The test method is verified by RRBS, ddPCR and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-2 | Human Low-Methylation Reference Standard | Average Methy <30% | The product is obtained by knocking out DNMT3A and DNMT3B proteins at the cellular level and extracting gDNA. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-3 | Human Medium-Methylation Reference Standard | 30% | This product is obtained by overexpressing DNMT3A and DNMT3B proteins in cells and extracting gDNA. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-4 | Human High-Methylation Reference Standard | Average Methy>70% | This product is a natural sample. The test method is verified by RRBS, WGBS and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
CBPE0390-5 | Human Hyper-Methylation Reference Standard | Average Methy≈100% | The whole genome methylated DNA of this product is obtained by modifying human genomic DNA with methyltransferase (M.SssI), which can be used as a positive control for DNA methylation analysis. The test method is verified by RRBS, ddPCR and enzyme digestion. The enzyme digestion method uses McrBc or MspI and HpaII to digest the sample. The sample is derived from a cell line and is closer to the real sample. |
Remark | 1. MspI can digest both unmethylated and methylated DNA, but is insensitive to methylation. 2. HpaII is sensitive to CpG methylation, but cannot digest methylated DNA, only unmethylated DNA. 3. This product uses the same cell line | ||
Storage Conditions | 2~8℃ | ||
Expiry | 36 months from the date of manufacture | ||
Representative Data

Figure 1. DNA methylation analysis by Reduced representation bisulfite sequencing (RRBS) on different samples.

Figure 2. Different samples digested by MspI or HapII.
Representative Data

Figure 1. DNA methylation analysis by Reduced representation bisulfite sequencing (RRBS) on different samples.

Figure 2. Different samples digested by MspI or HapII.