- Home
- Products
- Services
- Resources
- About Us
- News
- Contact

loading
CBP80001-1/2/3/4/5/6/7/8
CBP80001-1/2/3/4/5/6/7/8
| Availability: | |
|---|---|
Tumor Mutational Burden (TMB) serves as a pivotal biomarker for predicting cancer immunotherapy efficacy, reflecting the number of mutations per megabase in tumor exomes. Our Clinical-Grade tTMB Reference Standard (Cat. No. CBP80001-9) is a calibrated matrix reference material designed to standardize TMB measurement across next-generation sequencing (NGS) platforms, addressing the poor correlation among different assay systems in clinical settings .
Material form: Available in gDNA format (1µg per unit) with optional FFPE and ctDNA derivatives
TMB gradient range: Covers 2–28 mutations/Mb to mimic diverse cancer types
Validation basis: Verified via whole-exome sequencing (WES) and ddPCR for batch consistency
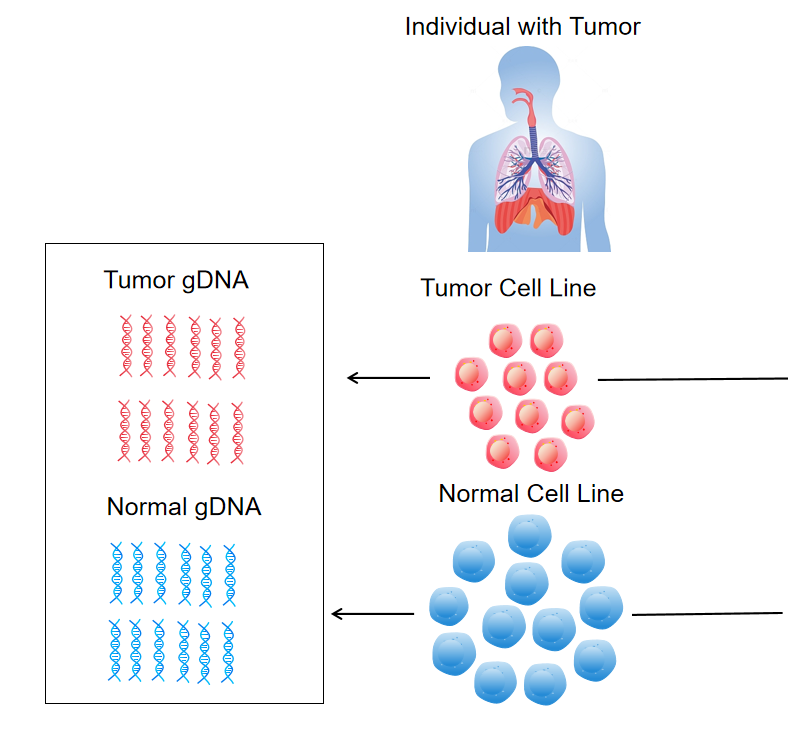
Cell line-derived matrix highly simulates patient samples, ensuring clinical relevance
ISO9001-certified production process guarantees traceability and minimal batch variation
Paired normal/tumor controls enable germline mutation filtering for accurate somatic variant calling
Compatible with targeted NGS panels and WES platforms commonly used in cancer I-O research
Adjustable tumor content ratios (10%–100%) simulate heterogeneous clinical samples
Stable under standard laboratory storage conditions with 12-month shelf life
Includes complete WES datasets and open-source TMB calculation pipelines
Provides detailed validation reports for regulatory submission (e.g., NMPA/FDA filings)
Optimize TMB detection workflows for IVD kit development
Validate analytical performance (sensitivity, specificity, linearity) of sequencing assays
Calibrate bioinformatics pipelines for consistent TMB scoring
Routine quality monitoring of clinical NGS platforms
Inter-laboratory proficiency testing for TMB measurement harmonization
Batch-to-batch verification of diagnostic reagents
Compare TMB results across different sequencing systems (Illumina, Ion Torrent)
Bridge discrepancies between targeted panels and WES-based TMB assessments
Store at -20°C upon receipt; avoid repeated freeze-thaw cycles. Diluted samples are stable for 72 hours at 4°C.
Yes. It meets CLSI guidelines for reference materials and supports analytical validation in clinical trials .
Custom gradients and matrix compositions (e.g., tissue-specific FFPE) are available upon request .
Each lot comes with certified TMB values calculated by WES (500X coverage) and cross-validated by ddPCR .
Manufactured in ISO9001-accredited laboratories in Nanjing and Shanghai
Rigorous multi-step validation involving 8+ independent testing centers
10+ years of expertise in molecular diagnostic reference standards
In-house WES and digital PCR platforms for real-time quality control
Dedicated technical support for assay optimization
Fast turnaround for custom orders and regulatory documentation assistance
General information
Cat.No. | ID | Format | Unit Size | TMB Value | Method | Buffer | Storage Conditions | Expiry |
CBP80001-1 | tTMB-P1 | Genomic DNA | 1ug+1ug | 5.37 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-2 | tTMB-P2 | Genomic DNA | 1ug+1ug | 9.84 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-3 | tTMB-P3 | Genomic DNA | 1ug+1ug | 12.41 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-4 | tTMB-P4 | Genomic DNA | 1ug+1ug | 21.09 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-5 | tTMB-P5 | Genomic DNA | 1ug+1ug | 27.15 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-6 | tTMB-P6 | Genomic DNA | 1ug+1ug | 8.98 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-7 | tTMB-P7 | Genomic DNA | 1ug+1ug | 6.83 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-8 | tTMB-P8 | Genomic DNA | 1ug+1ug | 27.15 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
Detection Methods
Detailed Data
1. tTMB-P1(5.37) Reference Standard CBP80001-1
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0153 | CBP80001-1N | B lymphoblast, Female | 500x WES | Same individual
|
DC198N0152 | CBP80001-1T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0 capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0152 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 180 | 172 | 168 | 166 |
TMB value | 5.37 | 5.13 | 5.01 | 4.95 |
2. tTMB-P2(9.84) Reference Standard CBP80001-2
| Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0159 | CBP80001-2N | B lymphoblast, male | 500x WES | Same individual
|
DC198N0158 | CBP80001-2T | Lung adenocarcinoma, male | 500x WES | |
IDT xGenExome Research Panel v1.0 capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0158 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 330 | 313 | 304 | 297 |
TMB value | 9.84 | 9.34 | 9.07 | 8.86 |
3. tTMB-P3(12.41) Reference Standard CBP80001-3
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0161 | CBP80001-3N | B lymphoblast, Female | 500x WES | Same individual
|
DC198N0160 | CBP80001-3T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0160 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 416 | 398 | 386 | 383 |
TMB value | 12.41 | 11.87 | 11.52 | 11.43 |
4. tTMB-P4(21.09) Reference Standard CBP80001-4
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0163 | CBP80001-4N | B lymphoblast, male | 500x WES | Same individual
|
DC198N0162 | CBP80001-4T | Lung large cell carcinoma, male | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0162 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 707 | 698 | 691 | 688 |
TMB value | 21.09 | 20.82 | 20.61 | 20.53 |
5. tTMB-P5(27.15) Reference Standard CBP80001-5
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0155 | CBP80001-5N | B lymphoblast, Female | 500x WES | Same individual
|
DC198N0154 | CBP80001-5T | stage 4, adenocarcinoma Lung, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0154 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 910 | 898.00 | 888 | 884 |
TMB value | 27.15 | 26.79 | 26.49 | 26.37 |
6. tTMB-P6(8.98) Reference Standard CBP80001-6
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0167 | CBP80001-6N | B lymphoblast, Female | 500x WES | Same individual
|
DC198N0166 | CBP80001-6T | Breast Cancer, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0166 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 301 | 275 | 259 | 254 |
TMB value | 8.98 | 8.20 | 7.73 | 7.58 |
7. tTMB-P7(6.83) Reference Standard CBP80001-7
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0157 | CBP80001-7N | B lymphoblast, Female | 500x WES | Same individual
|
DC198N0156 | CBP80001-7T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0156 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 229 | 218 | 208 | 201 |
TMB value | 6.83 | 6.50 | 6.21 | 6.00 |
8. tTMB-P8(27.15) Reference Standard CBP80001-8
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0165 | CBP80001-8N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0164 | CBP80001-8T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0164 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 910 | 840 | 766 | 685 |
TMB value | 27.15 | 25.06 | 22.85 | 20.44 |
Tumor Mutational Burden (TMB) serves as a pivotal biomarker for predicting cancer immunotherapy efficacy, reflecting the number of mutations per megabase in tumor exomes. Our Clinical-Grade tTMB Reference Standard (Cat. No. CBP80001-9) is a calibrated matrix reference material designed to standardize TMB measurement across next-generation sequencing (NGS) platforms, addressing the poor correlation among different assay systems in clinical settings .
Material form: Available in gDNA format (1µg per unit) with optional FFPE and ctDNA derivatives
TMB gradient range: Covers 2–28 mutations/Mb to mimic diverse cancer types
Validation basis: Verified via whole-exome sequencing (WES) and ddPCR for batch consistency

Cell line-derived matrix highly simulates patient samples, ensuring clinical relevance
ISO9001-certified production process guarantees traceability and minimal batch variation
Paired normal/tumor controls enable germline mutation filtering for accurate somatic variant calling
Compatible with targeted NGS panels and WES platforms commonly used in cancer I-O research
Adjustable tumor content ratios (10%–100%) simulate heterogeneous clinical samples
Stable under standard laboratory storage conditions with 12-month shelf life
Includes complete WES datasets and open-source TMB calculation pipelines
Provides detailed validation reports for regulatory submission (e.g., NMPA/FDA filings)
Optimize TMB detection workflows for IVD kit development
Validate analytical performance (sensitivity, specificity, linearity) of sequencing assays
Calibrate bioinformatics pipelines for consistent TMB scoring
Routine quality monitoring of clinical NGS platforms
Inter-laboratory proficiency testing for TMB measurement harmonization
Batch-to-batch verification of diagnostic reagents
Compare TMB results across different sequencing systems (Illumina, Ion Torrent)
Bridge discrepancies between targeted panels and WES-based TMB assessments
Store at -20°C upon receipt; avoid repeated freeze-thaw cycles. Diluted samples are stable for 72 hours at 4°C.
Yes. It meets CLSI guidelines for reference materials and supports analytical validation in clinical trials .
Custom gradients and matrix compositions (e.g., tissue-specific FFPE) are available upon request .
Each lot comes with certified TMB values calculated by WES (500X coverage) and cross-validated by ddPCR .
Manufactured in ISO9001-accredited laboratories in Nanjing and Shanghai
Rigorous multi-step validation involving 8+ independent testing centers
10+ years of expertise in molecular diagnostic reference standards
In-house WES and digital PCR platforms for real-time quality control
Dedicated technical support for assay optimization
Fast turnaround for custom orders and regulatory documentation assistance
General information
Cat.No. | ID | Format | Unit Size | TMB Value | Method | Buffer | Storage Conditions | Expiry |
CBP80001-1 | tTMB-P1 | Genomic DNA | 1ug+1ug | 5.37 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-2 | tTMB-P2 | Genomic DNA | 1ug+1ug | 9.84 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-3 | tTMB-P3 | Genomic DNA | 1ug+1ug | 12.41 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-4 | tTMB-P4 | Genomic DNA | 1ug+1ug | 21.09 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-5 | tTMB-P5 | Genomic DNA | 1ug+1ug | 27.15 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-6 | tTMB-P6 | Genomic DNA | 1ug+1ug | 8.98 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-7 | tTMB-P7 | Genomic DNA | 1ug+1ug | 6.83 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
CBP80001-8 | tTMB-P8 | Genomic DNA | 1ug+1ug | 27.15 | WES | Tris-EDTA | 2~8℃ | 36 months from the date of manufacture |
Detection Methods

Detailed Data
1. tTMB-P1(5.37) Reference Standard CBP80001-1
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0153 | CBP80001-1N | B lymphoblast, Female | 500x WES | Same individual
|
DC198N0152 | CBP80001-1T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0 capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0152 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 180 | 172 | 168 | 166 |
TMB value | 5.37 | 5.13 | 5.01 | 4.95 |
2. tTMB-P2(9.84) Reference Standard CBP80001-2
| Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0159 | CBP80001-2N | B lymphoblast, male | 500x WES | Same individual
|
DC198N0158 | CBP80001-2T | Lung adenocarcinoma, male | 500x WES | |
IDT xGenExome Research Panel v1.0 capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0158 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 330 | 313 | 304 | 297 |
TMB value | 9.84 | 9.34 | 9.07 | 8.86 |
3. tTMB-P3(12.41) Reference Standard CBP80001-3
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0161 | CBP80001-3N | B lymphoblast, Female | 500x WES | Same individual
|
DC198N0160 | CBP80001-3T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0160 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 416 | 398 | 386 | 383 |
TMB value | 12.41 | 11.87 | 11.52 | 11.43 |
4. tTMB-P4(21.09) Reference Standard CBP80001-4
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0163 | CBP80001-4N | B lymphoblast, male | 500x WES | Same individual
|
DC198N0162 | CBP80001-4T | Lung large cell carcinoma, male | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0162 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 707 | 698 | 691 | 688 |
TMB value | 21.09 | 20.82 | 20.61 | 20.53 |
5. tTMB-P5(27.15) Reference Standard CBP80001-5
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0155 | CBP80001-5N | B lymphoblast, Female | 500x WES | Same individual
|
DC198N0154 | CBP80001-5T | stage 4, adenocarcinoma Lung, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0154 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 910 | 898.00 | 888 | 884 |
TMB value | 27.15 | 26.79 | 26.49 | 26.37 |
6. tTMB-P6(8.98) Reference Standard CBP80001-6
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0167 | CBP80001-6N | B lymphoblast, Female | 500x WES | Same individual
|
DC198N0166 | CBP80001-6T | Breast Cancer, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0166 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 301 | 275 | 259 | 254 |
TMB value | 8.98 | 8.20 | 7.73 | 7.58 |
7. tTMB-P7(6.83) Reference Standard CBP80001-7
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0157 | CBP80001-7N | B lymphoblast, Female | 500x WES | Same individual
|
DC198N0156 | CBP80001-7T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0156 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 229 | 218 | 208 | 201 |
TMB value | 6.83 | 6.50 | 6.21 | 6.00 |
8. tTMB-P8(27.15) Reference Standard CBP80001-8
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0165 | CBP80001-8N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0164 | CBP80001-8T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0164 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 910 | 840 | 766 | 685 |
TMB value | 27.15 | 25.06 | 22.85 | 20.44 |
Detection Methods
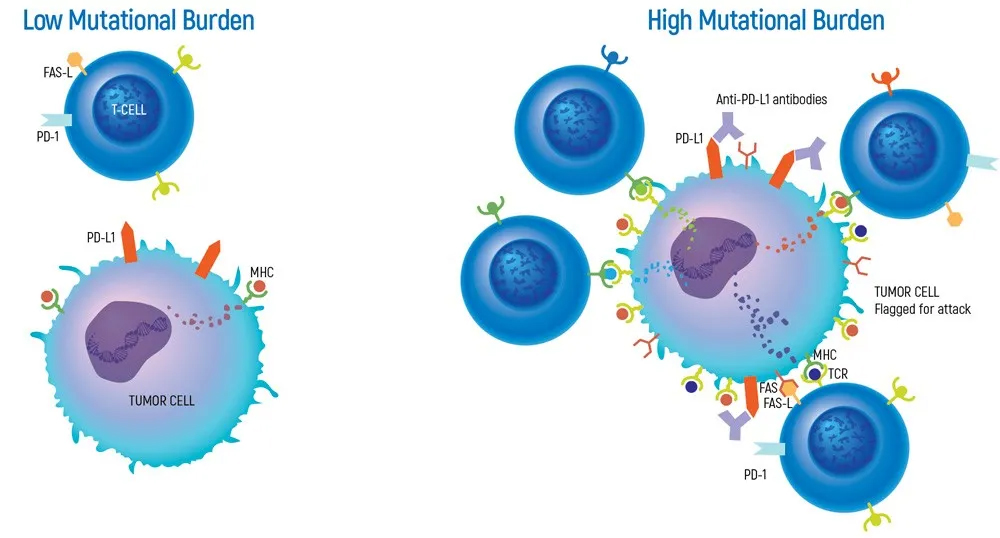
The current detection methods on the market refer to the number of somatic mutations per million bases (Mb) in the coding region of the patient's targeted sequencing, including point mutations and insertions and deletions. The number of somatic mutations in different cancers ranges from 0.01 mutations/Mb to more than 400 mutations/Mb.
The higher the tumor mutation load, the more likely the corresponding tumor-related carcinogenic mutations are, the more prominent the tumor's personality is, and the more different it is from normal cells.

Regarding TMB detection: Traditional detection technology is to analyze the patient's TMB by taking the patient's tumor tissue (tTMB). Currently, blood TMB (bTMB) can be detected, and compared with tissue detection, blood detection is more convenient and faster, and the non-invasive operation method also avoids more pain for patients.
Detection Methods
The current detection methods on the market refer to the number of somatic mutations per million bases (Mb) in the coding region of the patient's targeted sequencing, including point mutations and insertions and deletions. The number of somatic mutations in different cancers ranges from 0.01 mutations/Mb to more than 400 mutations/Mb.
The higher the tumor mutation load, the more likely the corresponding tumor-related carcinogenic mutations are, the more prominent the tumor's personality is, and the more different it is from normal cells.

Regarding TMB detection: Traditional detection technology is to analyze the patient's TMB by taking the patient's tumor tissue (tTMB). Currently, blood TMB (bTMB) can be detected, and compared with tissue detection, blood detection is more convenient and faster, and the non-invasive operation method also avoids more pain for patients.
Detailed Data
1. tTMB-P1(5.37) Reference Standard CBP80001-1
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0153 | CBP80001-1N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0152 | CBP80001-1T | Ductal breast carcinoma, Female | 500x WES | |
| IDT xGenExome Research Panel v1.0 capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0152 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 180 | 172 | 168 | 166 |
TMB value | 5.37 | 5.13 | 5.01 | 4.95 |
2. tTMB-P2(9.84) Reference Standard CBP80001-2
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0159 | CBP80001-2N | B lymphoblast, male | 500x WES | Same individual |
DC198N0158 | CBP80001-2T | Lung adenocarcinoma, male | 500x WES | |
IDT xGenExome Research Panel v1.0 capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0158 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 330 | 313 | 304 | 297 |
TMB value | 9.84 | 9.34 | 9.07 | 8.86 |
3. tTMB-P3(12.41) Reference Standard CBP80001-3
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0161 | CBP80001-3N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0160 | CBP80001-3T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0160 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 416 | 398 | 386 | 383 |
TMB value | 12.41 | 11.87 | 11.52 | 11.43 |
4. tTMB-P4(21.09) Reference Standard CBP80001-4
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0163 | CBP80001-4N | B lymphoblast, male | 500x WES | Same individual |
DC198N0162 | CBP80001-4T | Lung large cell carcinoma, male | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0162 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 707 | 698 | 691 | 688 |
TMB value | 21.09 | 20.82 | 20.61 | 20.53 |
5. tTMB-P5(27.15) Reference Standard CBP80001-5
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0155 | CBP80001-5N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0154 | CBP80001-5T | stage 4, adenocarcinoma Lung, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0154 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 910 | 898.00 | 888 | 884 |
TMB value | 27.15 | 26.79 | 26.49 | 26.37 |
6. tTMB-P6(8.98) Reference Standard CBP80001-6
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0167 | CBP80001-6N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0166 | CBP80001-6T | Breast Cancer, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0166 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 301 | 275 | 259 | 254 |
TMB value | 8.98 | 8.20 | 7.73 | 7.58 |
7. tTMB-P7(6.83) Reference Standard CBP80001-7
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0157 | CBP80001-7N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0156 | CBP80001-7T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0156 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 229 | 218 | 208 | 201 |
TMB value | 6.83 | 6.50 | 6.21 | 6.00 |
8. tTMB-P8(27.15) Reference Standard CBP80001-8
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0165 | CBP80001-8N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0164 | CBP80001-8T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0164 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 910 | 840 | 766 | 685 |
TMB value | 27.15 | 25.06 | 22.85 | 20.44 |
Detailed Data
1. tTMB-P1(5.37) Reference Standard CBP80001-1
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0153 | CBP80001-1N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0152 | CBP80001-1T | Ductal breast carcinoma, Female | 500x WES | |
| IDT xGenExome Research Panel v1.0 capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0152 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 180 | 172 | 168 | 166 |
TMB value | 5.37 | 5.13 | 5.01 | 4.95 |
2. tTMB-P2(9.84) Reference Standard CBP80001-2
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0159 | CBP80001-2N | B lymphoblast, male | 500x WES | Same individual |
DC198N0158 | CBP80001-2T | Lung adenocarcinoma, male | 500x WES | |
IDT xGenExome Research Panel v1.0 capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0158 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 330 | 313 | 304 | 297 |
TMB value | 9.84 | 9.34 | 9.07 | 8.86 |
3. tTMB-P3(12.41) Reference Standard CBP80001-3
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0161 | CBP80001-3N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0160 | CBP80001-3T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0160 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 416 | 398 | 386 | 383 |
TMB value | 12.41 | 11.87 | 11.52 | 11.43 |
4. tTMB-P4(21.09) Reference Standard CBP80001-4
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0163 | CBP80001-4N | B lymphoblast, male | 500x WES | Same individual |
DC198N0162 | CBP80001-4T | Lung large cell carcinoma, male | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0162 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 707 | 698 | 691 | 688 |
TMB value | 21.09 | 20.82 | 20.61 | 20.53 |
5. tTMB-P5(27.15) Reference Standard CBP80001-5
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0155 | CBP80001-5N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0154 | CBP80001-5T | stage 4, adenocarcinoma Lung, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0154 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 910 | 898.00 | 888 | 884 |
TMB value | 27.15 | 26.79 | 26.49 | 26.37 |
6. tTMB-P6(8.98) Reference Standard CBP80001-6
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0167 | CBP80001-6N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0166 | CBP80001-6T | Breast Cancer, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0166 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 301 | 275 | 259 | 254 |
TMB value | 8.98 | 8.20 | 7.73 | 7.58 |
7. tTMB-P7(6.83) Reference Standard CBP80001-7
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0157 | CBP80001-7N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0156 | CBP80001-7T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0156 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 229 | 218 | 208 | 201 |
TMB value | 6.83 | 6.50 | 6.21 | 6.00 |
8. tTMB-P8(27.15) Reference Standard CBP80001-8
Sample ID | Cat.No. | Background | Assay | Comments |
DC198N0165 | CBP80001-8N | B lymphoblast, Female | 500x WES | Same individual |
DC198N0164 | CBP80001-8T | Ductal breast carcinoma, Female | 500x WES | |
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M, CDS Region 33.52M; HiSeq X-TEN,af choose 0.02 cut off,Filter again. DC198N0164 is 100% Tumor sample. | ||||
Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 910 | 840 | 766 | 685 |
TMB value | 27.15 | 25.06 | 22.85 | 20.44 |