- Home
- Products
- Services
- Resources
- About Us
- News
- Contact

loading
CBPO0005-CBPO0007
CBPO0005-CBPO0007
| Availability: | |
|---|---|
Description
In the STR experiment, how to judge the accuracy of the results is essential. CB-Gene now introduces STR-related standards, which mainly include 1. Cell identification: autosomal series and sex chromosome series, as well as species pollution. 2.Reproductive inheritance: chromosome aneuploid pairing standard (mother and fetus) and maternal cell pollution detection standard.
The proportion is the quality proportion:10% mixed sample ,20% mixed sample and 30% mixed sample.
General information
Name | 10% STR Reference Standard | 20% STR Reference Standard | 30% STR Reference Standard |
Cat. No. | CBPO0005 | CBPO0006 | CBPO0007 |
Format | Genomic DNA | Genomic DNA | Genomic DNA |
Unit Size | 1ug | 1ug | 1ug |
Buffer | TE Buffer | TE Buffer | TE Buffer |
Concentration | Download for COA | Download for COA | Download for COA |
Purofication | Download for COA | Download for COA | Download for COA |
DNA Electrophoresis | Download for COA | Download for COA | Download for COA |
Storage Conditions | 2~8℃ | 2~8℃ | 2~8℃ |
Expiry | 36 months from the date of manufacture | 36 months from the date of manufacture | 36 months from the date of manufacture |
Technical Data
| Sample A | Sample B | |||||||
| CBPO0005 | 10% STR typing | 90% STR typing | ||||||
| CBPO0006 | 20% STR typing | 80% STR typing | ||||||
| CBPO0007 | 30% STR typing | 70% STR typing | ||||||
| Data | Allele | Data | Allele | Data | Allele | Data | Allele | Data |
CSF1PO | 10,12 | D18S51 | 15,19 | CSF1PO | 10,11, or | D18S51 | 15,18 | |
10,11,12* | ||||||||
D2S1338 | 19,23 | D19S433 | 14,15 | D2S1338 | 23,23 | D19S433 | 13,14 | |
D2S441 | 10,14 | D21S11 | 30,30 | D2S441 | 11,12 | D21S11 | 29,30 | |
D3S1358 | 14,15 | FGA | 23,24 | D3S1358 | 15,17 | FGA | 24,26 | |
D5S818 | 11,11 | Penta D | 12,12 | D5S818 | 11,13 | Penta D | 8,12 | |
D6S1043 | 12,18 | Penta E | 12,13 | D6S1043 | 12,12 | Penta E | 11,11 | |
D7S820 | 10,11 | TH01 | 8,9.3 | D7S820 | 11,11 | TH01 | 6,9.3 | |
D8S1179 | 13,13 | TPOX | 8,8 | D8S1179 | 12,13 | TPOX | 8,9 | |
D12S391 | 18,20 | vWA | 17,18 | D12S391 | 18,24 | vWA | 17,17 | |
D13S317 | 11,11 | Amel | X,X | D13S317 | 11,11 | Amel | X,Y | |
D16S539 | 11,12 | D16S539 | 11,11 | |||||
Remark*: The relative intensity of the 12 allele is less than 10 % of the dominant 10 allele.
Product Workflow
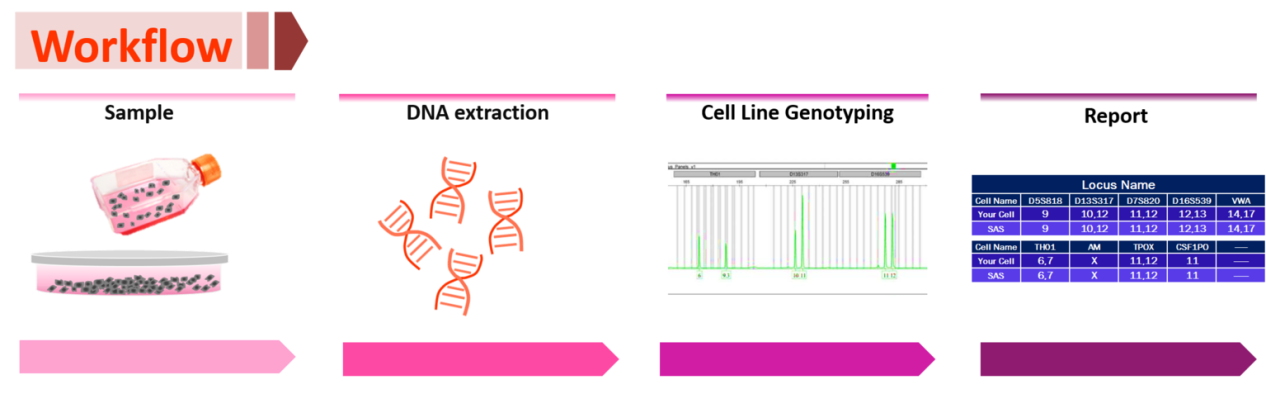
Product development
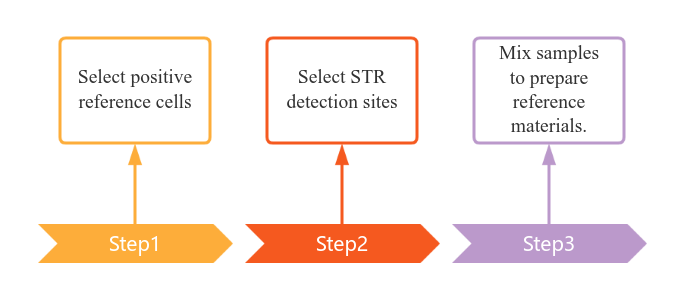
Case Showcase
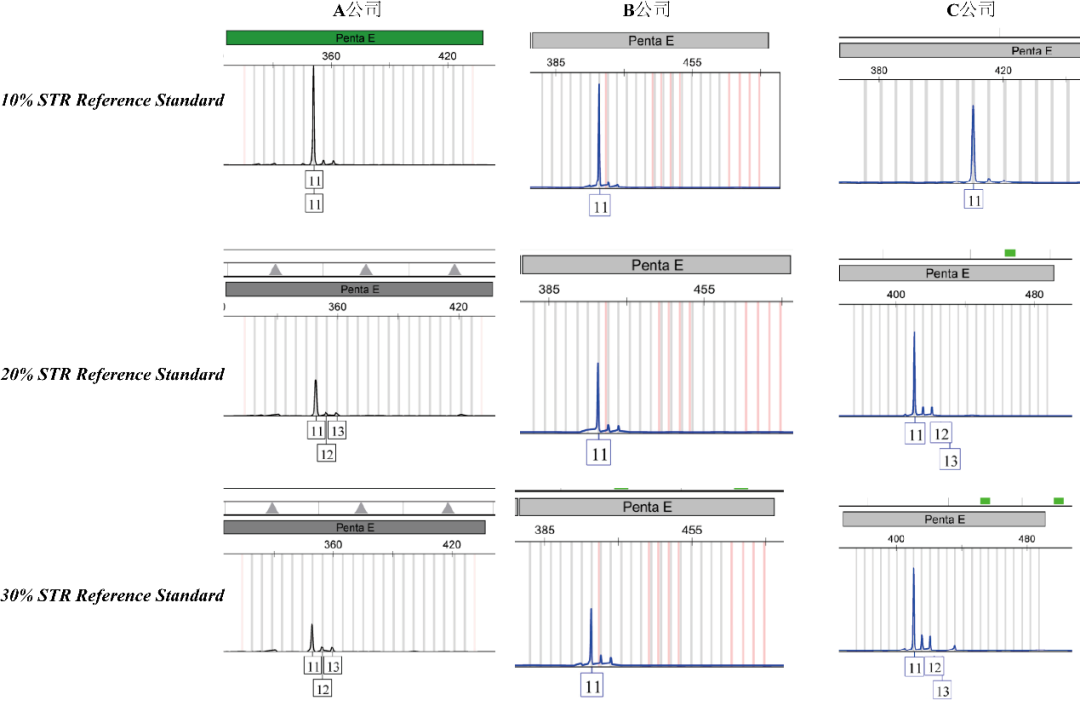
CB-Gene accurately prepare mixed gDNA samples by dPCR with different proportions of cell contamination: 10% contaminated samples, 20% contaminated samples and 30% contaminated samples, and sent the above samples to three testing companies (marked as A, B, and C) and tested them using their CE method
Description
In the STR experiment, how to judge the accuracy of the results is essential. CB-Gene now introduces STR-related standards, which mainly include 1. Cell identification: autosomal series and sex chromosome series, as well as species pollution. 2.Reproductive inheritance: chromosome aneuploid pairing standard (mother and fetus) and maternal cell pollution detection standard.
The proportion is the quality proportion:10% mixed sample ,20% mixed sample and 30% mixed sample.
General information
Name | 10% STR Reference Standard | 20% STR Reference Standard | 30% STR Reference Standard |
Cat. No. | CBPO0005 | CBPO0006 | CBPO0007 |
Format | Genomic DNA | Genomic DNA | Genomic DNA |
Unit Size | 1ug | 1ug | 1ug |
Buffer | TE Buffer | TE Buffer | TE Buffer |
Concentration | Download for COA | Download for COA | Download for COA |
Purofication | Download for COA | Download for COA | Download for COA |
DNA Electrophoresis | Download for COA | Download for COA | Download for COA |
Storage Conditions | 2~8℃ | 2~8℃ | 2~8℃ |
Expiry | 36 months from the date of manufacture | 36 months from the date of manufacture | 36 months from the date of manufacture |
Technical Data
| Sample A | Sample B | |||||||
| CBPO0005 | 10% STR typing | 90% STR typing | ||||||
| CBPO0006 | 20% STR typing | 80% STR typing | ||||||
| CBPO0007 | 30% STR typing | 70% STR typing | ||||||
| Data | Allele | Data | Allele | Data | Allele | Data | Allele | Data |
CSF1PO | 10,12 | D18S51 | 15,19 | CSF1PO | 10,11, or | D18S51 | 15,18 | |
10,11,12* | ||||||||
D2S1338 | 19,23 | D19S433 | 14,15 | D2S1338 | 23,23 | D19S433 | 13,14 | |
D2S441 | 10,14 | D21S11 | 30,30 | D2S441 | 11,12 | D21S11 | 29,30 | |
D3S1358 | 14,15 | FGA | 23,24 | D3S1358 | 15,17 | FGA | 24,26 | |
D5S818 | 11,11 | Penta D | 12,12 | D5S818 | 11,13 | Penta D | 8,12 | |
D6S1043 | 12,18 | Penta E | 12,13 | D6S1043 | 12,12 | Penta E | 11,11 | |
D7S820 | 10,11 | TH01 | 8,9.3 | D7S820 | 11,11 | TH01 | 6,9.3 | |
D8S1179 | 13,13 | TPOX | 8,8 | D8S1179 | 12,13 | TPOX | 8,9 | |
D12S391 | 18,20 | vWA | 17,18 | D12S391 | 18,24 | vWA | 17,17 | |
D13S317 | 11,11 | Amel | X,X | D13S317 | 11,11 | Amel | X,Y | |
D16S539 | 11,12 | D16S539 | 11,11 | |||||
Remark*: The relative intensity of the 12 allele is less than 10 % of the dominant 10 allele.
Product Workflow

Product development

Case Showcase
CB-Gene accurately prepare mixed gDNA samples by dPCR with different proportions of cell contamination: 10% contaminated samples, 20% contaminated samples and 30% contaminated samples, and sent the above samples to three testing companies (marked as A, B, and C) and tested them using their CE method

Technical Data
| Sample A | Sample B | |||||||
| CBPO0005 | 10% STR typing | 90% STR typing | ||||||
| CBPO0006 | 20% STR typing | 80% STR typing | ||||||
| CBPO0007 | 30% STR typing | 70% STR typing | ||||||
| Data | Allele | Data | Allele | Data | Allele | Data | Allele | Data |
CSF1PO | 10,12 | D18S51 | 15,19 | CSF1PO | 10,11, or | D18S51 | 15,18 | |
10,11,12* | ||||||||
D2S1338 | 19,23 | D19S433 | 14,15 | D2S1338 | 23,23 | D19S433 | 13,14 | |
D2S441 | 10,14 | D21S11 | 30,30 | D2S441 | 11,12 | D21S11 | 29,30 | |
D3S1358 | 14,15 | FGA | 23,24 | D3S1358 | 15,17 | FGA | 24,26 | |
D5S818 | 11,11 | Penta D | 12,12 | D5S818 | 11,13 | Penta D | 8,12 | |
D6S1043 | 12,18 | Penta E | 12,13 | D6S1043 | 12,12 | Penta E | 11,11 | |
D7S820 | 10,11 | TH01 | 8,9.3 | D7S820 | 11,11 | TH01 | 6,9.3 | |
D8S1179 | 13,13 | TPOX | 8,8 | D8S1179 | 12,13 | TPOX | 8,9 | |
D12S391 | 18,20 | vWA | 17,18 | D12S391 | 18,24 | vWA | 17,17 | |
D13S317 | 11,11 | Amel | X,X | D13S317 | 11,11 | Amel | X,Y | |
D16S539 | 11,12 | D16S539 | 11,11 | |||||
Remark*: The relative intensity of the 12 allele is less than 10 % of the dominant 10 allele.
Technical Data
| Sample A | Sample B | |||||||
| CBPO0005 | 10% STR typing | 90% STR typing | ||||||
| CBPO0006 | 20% STR typing | 80% STR typing | ||||||
| CBPO0007 | 30% STR typing | 70% STR typing | ||||||
| Data | Allele | Data | Allele | Data | Allele | Data | Allele | Data |
CSF1PO | 10,12 | D18S51 | 15,19 | CSF1PO | 10,11, or | D18S51 | 15,18 | |
10,11,12* | ||||||||
D2S1338 | 19,23 | D19S433 | 14,15 | D2S1338 | 23,23 | D19S433 | 13,14 | |
D2S441 | 10,14 | D21S11 | 30,30 | D2S441 | 11,12 | D21S11 | 29,30 | |
D3S1358 | 14,15 | FGA | 23,24 | D3S1358 | 15,17 | FGA | 24,26 | |
D5S818 | 11,11 | Penta D | 12,12 | D5S818 | 11,13 | Penta D | 8,12 | |
D6S1043 | 12,18 | Penta E | 12,13 | D6S1043 | 12,12 | Penta E | 11,11 | |
D7S820 | 10,11 | TH01 | 8,9.3 | D7S820 | 11,11 | TH01 | 6,9.3 | |
D8S1179 | 13,13 | TPOX | 8,8 | D8S1179 | 12,13 | TPOX | 8,9 | |
D12S391 | 18,20 | vWA | 17,18 | D12S391 | 18,24 | vWA | 17,17 | |
D13S317 | 11,11 | Amel | X,X | D13S317 | 11,11 | Amel | X,Y | |
D16S539 | 11,12 | D16S539 | 11,11 | |||||
Remark*: The relative intensity of the 12 allele is less than 10 % of the dominant 10 allele.
Product Workflow

Product Workflow

Product Development

Product Development

Case Showcase
CB-Gene accurately prepare mixed gDNA samples by dPCR with different proportions of cell contamination: 10% contaminated samples, 20% contaminated samples and 30% contaminated samples, and sent the above samples to three testing companies (marked as A, B, and C) and tested them using their CE method

Case Showcase
CB-Gene accurately prepare mixed gDNA samples by dPCR with different proportions of cell contamination: 10% contaminated samples, 20% contaminated samples and 30% contaminated samples, and sent the above samples to three testing companies (marked as A, B, and C) and tested them using their CE method
