- Home
- Products
- Services
- Resources
- About Us
- News
- Contact

Views: 0 Author: Site Editor Publish Time: 2025-04-21 Origin: Site
Background
Non-invasive prenatal testing (NIPT) mainly uses NGS to detect free DNA in the peripheral blood of pregnant women (free DNA refers to DNA that has been fragmented under natural conditions in plasma) to detect whether the fetus has chromosomal aneuploidy. Common chromosomal diseases include autosomal aneuploidy, sex chromosome aneuploidy, and chromosome copy number variation. A small number of people with chromosomal aberrations can survive to birth, often causing multiple malformations, mental retardation, growth retardation, and multi-system dysfunction. At present, prenatal screening and prenatal diagnosis are the premise and main measures to avoid the birth of children with fatal and disabling chromosomal diseases.
Common screening methods
Common methods for screening chromosomal abnormalities include G-banding, fluorescence in situ hybridization (FISH), DNA microarray (CMA), NIPT, NIPT-Plus, CNV-seq, etc.
Chromosome karyotype analysis (G-banding) is generally used to detect the number and large structural abnormalities of chromosomes, and is the gold standard for detection. FISH can theoretically detect information at any position in the genome, but it requires corresponding probes. Currently, the commonly used clinical test kits are for chromosomes 13, 18, 21 and sex chromosomes. NIPT and NIPT- Plus are based on NGS methods. NIPT uses Z scores to determine whether the chromosomes are abnormal. Non-invasive prenatal testing for single-gene diseases is for single-gene genetic diseases. DNA microarray mainly detects microdeletions and microduplications of chromosomes, and uses log R ratio to determine whether they are abnormal.
Detection method | Detection range | Disadvantages |
Chromosome karyotype analysis (G banding) | Analyze abnormalities in chromosome structure, loss of large chromosome fragments, duplications and abnormal positions | Cannot detect small (<5M) genomic variations |
DNA microarray (CMA) | Abnormal chromosome number and structure | Can only detect known abnormal chromosome positions |
DNA microarray (CMA) | Chromosome copy number variation, as well as chromosome microdeletion, microduplication, uniparental disomy | Cannot detect gene level, mainly for some common chromosomal abnormalities |
Non-invasive single gene disease detection | Monogenic genetic disease | Highly targeted, for a single disease |
NIPT (NGS) | Abnormal chromosome number | Cannot detect chromosome microdeletion/microduplication NIPT-plus (NGS) |
NIPT-plus (NGS) | Chromosome copy number variation, as well as chromosome microdeletions and microduplications | Can only be used as a screening method, not as a diagnostic method, and cannot detect genetic levels |
CNV-Seq | Whole genome detection, high sensitivity, high precision | Data processing is complex, and sample processing standards are high |
Down syndrome screening | Can only detect trisomy 18, trisomy 21 and neural tube defects | Limited detection range, low detection rate, and a certain probability of false positives |
Table 1. Comparison of common chromosome detection methods
In the process of detection, quality control products and reference products are indispensable. For NIPT detection, CB-Gene has launched chromosome aneuploidy reference products and quality control products, including autosomes and sex chromosomes, as well as microdeletion and microduplication quality control products and reference products. In the future, chimera reference products will be launched one after another.
Partial product data display
Product Name | Chromosomal abnormalities | Catalog ID | |
Common and Rare chromosomal aneuploidy standards | Trisomy 21(47,XX,+21) Reference Standard | Chromosome 21 trisomy standard, female | CBPJ0001 |
Trisomy 21 (47,XY,+21) Reference Standard | Chromosome 21 trisomy standard, male | CBPJ0009 | |
Trisomy 18(47,XX,+18) Reference Standard | Chromosome 18 trisomy standard, female | CBPJ0002 | |
Trisomy 13 (47,XY,+13) Reference Standard | Chromosome 13 trisomy standard, female | CBPJ0010 | |
Trisomy 9(47,XY,+9)Reference Standard | Chromosome 9 trisomy standard, male | CBPJ0014 | |
Sex chromosome aneuploidy standards | Klinefelter Syndrome (47,XXY) Reference Standard | X chromosome non-global ploidy standard | CBPJ0005 |
Microdeletion and microduplication standards | Trisomy 9 (47,XY,+9, Gain 9p24.3p13.1) Reference Standard | 9q11 microdeletion standard, male | CBPJ0003 |
Angelman syndrome (46,XX,del(15)(q11q13)) Reference Standard | Angelman Syndrome Standards, Female | CBPJ0006 | |
Prader-Willi syndrome (46,XY,del(15)(q11.2q13)) Reference Standard | Prader-Willi syndrome standard, male | CBPJ0007 | |
18P-syndrome (46,XX,del(18)(p11.2)) Reference Standard | 18P-syndrome standard, female | CBPJ0008 | |
DiGeorge syndrome (46,XX,del(22)(q11)) Reference Standard | DiGeorge syndrome standard, female | CBPJ0011 | |
18Q-syndrome (46,XX,del(18)(q22)) Reference Standard | 18Q-syndrome standard, female | CBPJ0013 | |
11q23.3 del (46,XX,del(11)(q23.3)) Reference Standard | 11q23.3 del standard, female | CBPJ0015 | |
Negative control | Normal Karyotype (46,XY) Reference Standard | CBPJ0004 |
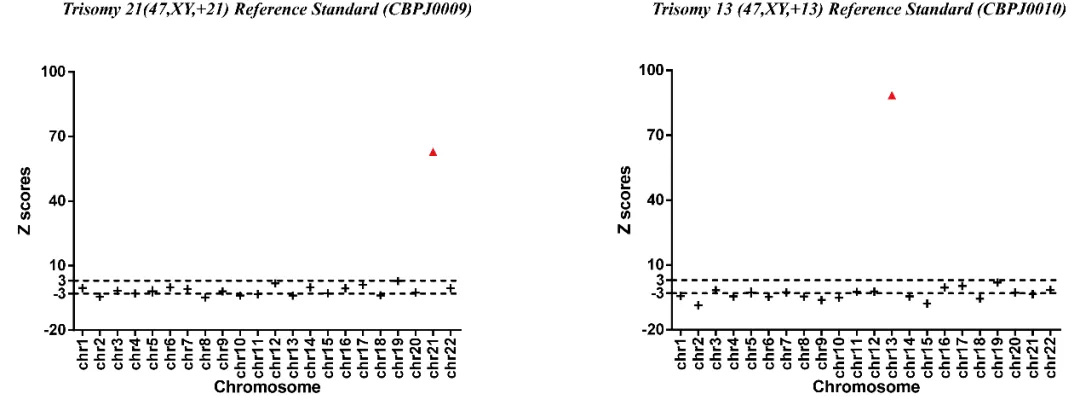
Fig 1. Results of NGS with Trisomy 21 (47,XY,+21) Reference Standard and Trisomy 13 (47,XY,+13) Reference Standard.
NGS technology can sequence cfDNA, and combined with information analysis methods, the risk rate of fetal chromosomal aneuploidy can be evaluated through Z scores. When Z scores are >3 or <-3, the chromosome is considered abnormal. Z scores >3 determine that the chromosome is duplicated, and Z scores <-3 determine that the chromosome is missing.
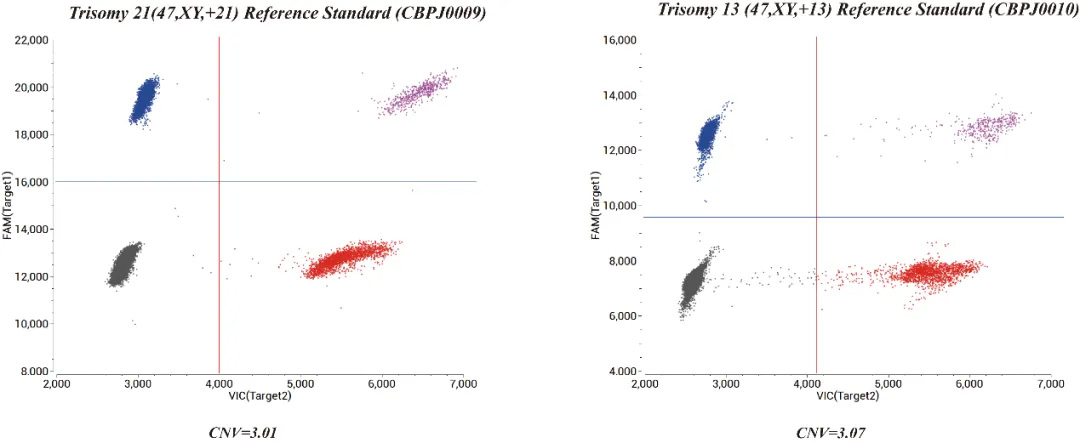
Fig 2. Results of ddPCR with Trisomy 21 (47,XY,+21) Reference Standard and Trisomy 13 (47,XY,+13) Reference Standard.
After determining the abnormal chromosome according to the NGS results, the primers and probes of the related abnormal chromosomes are designed according to the human genome sequence. The ddPCR method is verified again, and the chromosome situation is judged by the CNV value (by default, an amplification region is selected every interval of the entire chromosome).
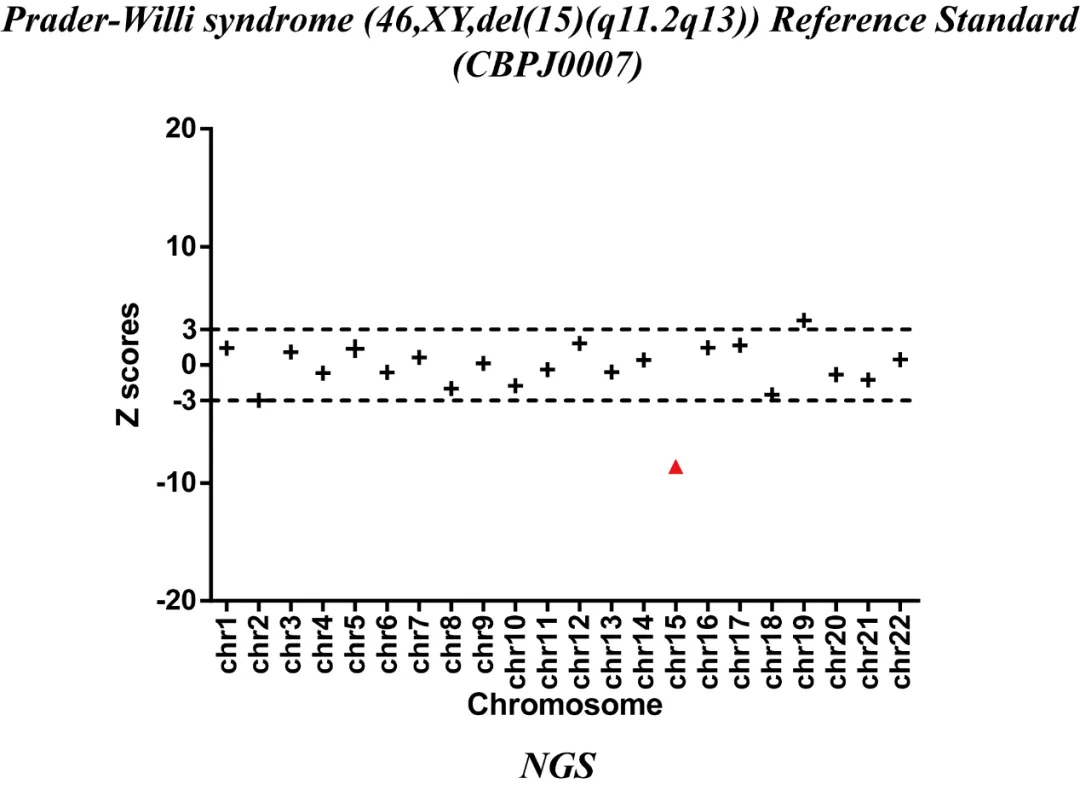
Fig 3. Results of NGS with Prader-Willi syndrome (46,XY,del(15)(q11.2q13)) Reference Standard.
Microdeletion and microduplication quality control products and reference products were verified by NGS and CMA methods. NGS can determine that chromosome 15 is deleted, and CMA method can determine that the microdeletion area of the sample is 15q11.2q13. After the deletion or duplication area is determined according to the above verification method, primers and probes are designed according to the deletion or duplication area, and ddPCR detection is performed to verify the abnormality of the sample again.
All samples are later simulated by enzyme digestion or ultrasound to simulate clinical samples. After DNA is fragmented, the reference products are accurately calibrated by ddPCR technology to accurately detect the proportion of fetal free DNA in the reference products. The samples tested by ddPCR are mixed with DNA-free plasma, which is closer to clinical samples.
The NIPT series reference products launched by CB-Gene can not only be applied to a variety of detection methods, but also can be used to evaluate whether the concentration of fetal free DNA determined by high-throughput sequencing is accurate.
This category is empty.