- Home
- Products
- Services
- Resources
- About Us
- News
- Contact

loading
CBP80001-9
CBP80001-9
| Availability: | |
|---|---|
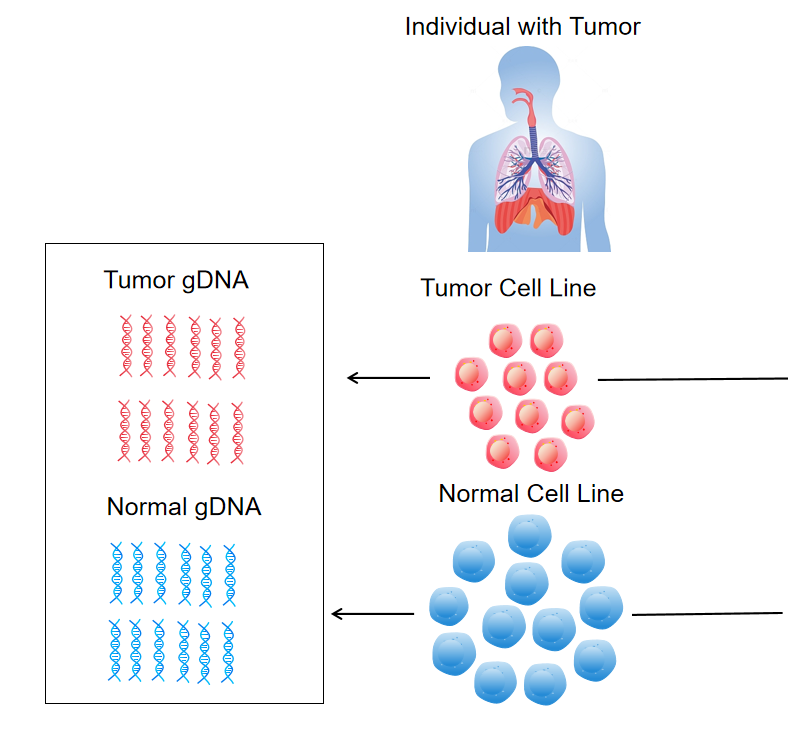
The Genomic DNA tTMB-P9 Standard is a specialized matrix reference standard developed by Cobio Biotech, serving as the 9th product in the company’s comprehensive tTMB (tissue Tumor Mutation Burden) standard series (complementing tTMB-P1 to tTMB-P8). Unlike single-tumor-content standards in the series, this product is uniquely designed to simulate the heterogeneity of clinical tissue samples, featuring four precisely calibrated tumor-normal (T-N) mixing ratios: 10% tumor (T) + 90% normal (N), 5%T + 95%N, 2%T + 98%N, and 1%T + 99%N.
The standard is derived from high-quality genomic DNA (gDNA) samples: the tumor component is isolated from a stage 4 lung adenocarcinoma patient’s tissue, while the normal component comes from B lymphoblasts of the same individual—ensuring genetic consistency and eliminating inter-individual background interference. For validation, the product undergoes 500× whole-exome sequencing (WES) using the IDT xGenExome Research Panel v1.0, which targets a 39 Mb coding region (CDS region: 33.52 Mb; Probe region: 51 Mb) and is sequenced on the HiSeq X-TEN platform. Post-sequencing, variants are filtered with allele frequency (AF) cutoffs ranging from 1% to 5%, generating reliable TMB values for each mixing ratio (e.g., 19.72 mutations/Mb at 10%T with 2% AF cutoff, 10.5 mutations/Mb at 5%T with 2% AF cutoff).
Its core function lies in quality control (QC) for NGS-based TMB detection workflows, particularly focusing on evaluating how tumor content variability impacts TMB measurement accuracy—a critical challenge in clinical practice where tissue samples often have low or uneven tumor cellularity.
Mimics real-world clinical scenarios where tissue samples rarely have 100% tumor content. The 1%–10% tumor content gradient covers common low-to-moderate tumor proportion cases (e.g., needle biopsy samples with high stromal content), allowing labs to test TMB assay performance under clinically relevant conditions.
Eliminates the limitation of single-tumor-content standards, which cannot reflect how low tumor cellularity affects mutation detection sensitivity.
Supports AF cutoff analysis across 1%–5%, a key parameter in TMB calculation. Labs can use the standard to determine the optimal AF threshold for their workflows and verify if the assay can consistently detect mutations at different cutoff levels.
Enables quantitative assessment of detection limits (LOD) and sensitivity: for example, the 1%T + 99%N ratio helps identify if an assay can reliably measure TMB in samples with extremely low tumor content (TMB value: 0.95 mutations/Mb at 2% AF cutoff).
Validated via 500× WES, a benchmark technology for TMB measurement, ensuring the standard’s TMB values are accurate and traceable. The IDT xGenExome Panel used aligns with common clinical exome sequencing protocols, enhancing compatibility with lab workflows.
Undergoes strict post-sequencing filtering (e.g., removing germline variants, low-quality reads) to ensure only high-confidence somatic mutations are counted, matching clinical TMB analysis pipelines.
Optimized for Illumina-based platforms (e.g., HiSeq X-TEN, as used in validation) and compatible with other NGS systems that support exome or targeted panel sequencing of the 39 Mb coding region. No additional protocol modifications are required for integration into existing workflows.
Used by kit manufacturers to validate the performance of new tTMB detection kits. During development, the standard can be spiked into normal gDNA to test if the kit maintains accuracy across different tumor content levels (e.g., confirming the kit can correctly measure TMB in 5%T samples without false lows).
Aids in setting kit specifications, such as defining the minimum tumor content required for reliable results (e.g., using the 2%T ratio to determine if the kit’s LOD meets clinical needs).
Clinical labs can use the standard to assess the stability of in-house TMB assays. For instance, testing the same batch of tTMB-P9 across multiple runs to verify inter-run reproducibility of TMB values at each tumor content ratio.
Supports comparison of different assay versions (e.g., updated probe panels or bioinformatics pipelines) by using the standard as a consistent reference to measure improvements in sensitivity or accuracy.
Serves as a positive control in daily TMB testing. Labs can run the standard alongside patient samples to monitor if the NGS workflow (e.g., DNA extraction, library preparation, sequencing) is functioning properly—if the standard’s TMB value deviates from the reference range, it indicates potential errors in the workflow.
Facilitates compliance with regulatory requirements: the standard provides traceable QC data, which is essential for meeting clinical laboratory accreditation standards (e.g., CAP, CLIA) for TMB testing.
The tTMB-P1 to tTMB-P8 standards are single-tumor-content (100%T) references with fixed TMB values (ranging from 5.37 to 27.15 mutations/Mb), designed primarily for calibrating TMB value ranges and verifying assay linearity. In contrast, tTMB-P9 is a matrix standard focused on tumor content variability (1%–10%T), targeting the evaluation of how low tumor cellularity impacts TMB measurement. The two types complement each other: P1-P8 ensure accurate TMB value assignment, while P9 ensures assay robustness in heterogeneous clinical samples.
No. The tTMB-P9 Standard is formulated with genomic DNA from tissue samples (gDNA) and is specifically designed for tissue TMB (tTMB) assay QC. For blood TMB (bTMB) testing, which uses circulating tumor DNA (ctDNA), Cobio Biotech offers dedicated bTMB standards (e.g., bTMB-P1), which are optimized for ctDNA’s unique characteristics (e.g., low concentration, fragmentation). Using tTMB-P9 for bTMB QC will not reflect the performance of ctDNA-specific workflows.
When stored at -20°C (avoiding repeated freeze-thaw cycles), the standard maintains stability for 12 months from the date of receipt. For short-term use (up to 1 month), it can be stored at 4°C. It is recommended to aliquot the standard into small volumes upon receipt to prevent degradation from repeated freeze-thaws.
The 1%T + 99%N ratio is the most challenging for TMB assays, as it contains extremely low levels of tumor-derived mutations. A reliable assay should detect a TMB value close to the reference (0.95 mutations/Mb at 2% AF cutoff). If the measured TMB is significantly lower than the reference, it indicates the assay may lack sufficient sensitivity for low-tumor-content samples. If the value is higher, it may suggest false-positive variant calls (e.g., from normal cell mutations or sequencing errors), requiring optimization of bioinformatics filters.
Every batch of tTMB-P9 Standard undergoes duplicate WES validation to confirm TMB value consistency (variation ≤ 5% from the reference range). Additionally, we test for DNA integrity (using agarose gel electrophoresis) and purity (A260/A280 ratio 1.8–2.0) to ensure the standard meets high-quality gDNA requirements—critical for reliable NGS performance.
The 1%–10% tumor content gradient is based on analysis of thousands of clinical tissue samples, addressing the most common issue labs face: inaccurate TMB results from low-tumor-content biopsies. By using this standard, labs can proactively identify and resolve workflow limitations before they affect patient test results.
We provide a detailed technical datasheet with reference TMB values for all tumor content ratios and AF cutoffs, as well as a recommended workflow for using the standard in QC. Our team of bioinformatics and molecular biology experts is also available to assist with data interpretation, assay optimization, and troubleshooting—ensuring labs get the most value from the product.
The tTMB-P9 Standard is developed in line with global regulatory guidelines for in vitro diagnostic (IVD) QC standards (e.g., FDA, NMPA). It has been used to support the development of NMPA-approved TMB detection kits (e.g., non-small cell lung cancer TMB kits), demonstrating its suitability for regulatory-compliant clinical workflows.
Name | tTMB-P9(matrix) Reference Standard |
Cat. No. | CBP80001-9 |
Format | Genomic DNA |
Size | 1ug+1ug |
Inventory Status | In Stock |
Buffer | Tris-EDTA |
Storage Conditions | 2~8℃ |
Expiry | 36 months from the date of manufacture |
Sample ID | Description | Cut off=1% | Cut off=2% | Cut off=3% | Cut off=4% | Cut off=5% |
DC208D0421 | 10%T+90%N | 22.94 | 19.72 | 16.53 | 12.50 | 8.41 |
DC208D0422 | 5%T+95%N | 15.75 | 10.50 | 5.82 | 2.92 | 1.37 |
DC208D0423 | 2%T+98%N | 8.29 | 2.12 | 0.75 | 0.42 | 0.21 |
DC208D0424 | 1%T+99%N | 4.15 | 0.95 | 0.27 | 0.18 | 0.09 |
500xWES; IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M; HiSeq X-TEN,af choose 0.01 cut off,call mutation,Filter again, and then calculate TMB according to the gradient cut off. Tumor sample: stage 4, adenocarcinoma Lung, Female Normal sample: B lymphoblast from the same individual | ||||||
Cat.No. | ID | TMB Value | Method |
CBP80001-1 | tTMB-P1 | 5.37 | WES |
CBP80001-2 | tTMB-P2 | 9.84 | WES |
CBP80001-3 | tTMB-P3 | 12.41 | WES |
CBP80001-4 | tTMB-P4 | 21.09 | WES |
CBP80001-5 | tTMB-P5 | 27.15 | WES |
CBP80001-6 | tTMB-P6 | 8.98 | WES |
CBP80001-7 | tTMB-P7 | 6.83 | WES |
CBP80001-8 | tTMB-P8 | 27.15 | WES |
CBP80001-10 | bTMB-P1 | 22.91 | WES |
The Genomic DNA tTMB-P9 Standard is a specialized matrix reference standard developed by Cobio Biotech, serving as the 9th product in the company’s comprehensive tTMB (tissue Tumor Mutation Burden) standard series (complementing tTMB-P1 to tTMB-P8). Unlike single-tumor-content standards in the series, this product is uniquely designed to simulate the heterogeneity of clinical tissue samples, featuring four precisely calibrated tumor-normal (T-N) mixing ratios: 10% tumor (T) + 90% normal (N), 5%T + 95%N, 2%T + 98%N, and 1%T + 99%N.
The standard is derived from high-quality genomic DNA (gDNA) samples: the tumor component is isolated from a stage 4 lung adenocarcinoma patient’s tissue, while the normal component comes from B lymphoblasts of the same individual—ensuring genetic consistency and eliminating inter-individual background interference. For validation, the product undergoes 500× whole-exome sequencing (WES) using the IDT xGenExome Research Panel v1.0, which targets a 39 Mb coding region (CDS region: 33.52 Mb; Probe region: 51 Mb) and is sequenced on the HiSeq X-TEN platform. Post-sequencing, variants are filtered with allele frequency (AF) cutoffs ranging from 1% to 5%, generating reliable TMB values for each mixing ratio (e.g., 19.72 mutations/Mb at 10%T with 2% AF cutoff, 10.5 mutations/Mb at 5%T with 2% AF cutoff).
Its core function lies in quality control (QC) for NGS-based TMB detection workflows, particularly focusing on evaluating how tumor content variability impacts TMB measurement accuracy—a critical challenge in clinical practice where tissue samples often have low or uneven tumor cellularity.

Mimics real-world clinical scenarios where tissue samples rarely have 100% tumor content. The 1%–10% tumor content gradient covers common low-to-moderate tumor proportion cases (e.g., needle biopsy samples with high stromal content), allowing labs to test TMB assay performance under clinically relevant conditions.
Eliminates the limitation of single-tumor-content standards, which cannot reflect how low tumor cellularity affects mutation detection sensitivity.
Supports AF cutoff analysis across 1%–5%, a key parameter in TMB calculation. Labs can use the standard to determine the optimal AF threshold for their workflows and verify if the assay can consistently detect mutations at different cutoff levels.
Enables quantitative assessment of detection limits (LOD) and sensitivity: for example, the 1%T + 99%N ratio helps identify if an assay can reliably measure TMB in samples with extremely low tumor content (TMB value: 0.95 mutations/Mb at 2% AF cutoff).
Validated via 500× WES, a benchmark technology for TMB measurement, ensuring the standard’s TMB values are accurate and traceable. The IDT xGenExome Panel used aligns with common clinical exome sequencing protocols, enhancing compatibility with lab workflows.
Undergoes strict post-sequencing filtering (e.g., removing germline variants, low-quality reads) to ensure only high-confidence somatic mutations are counted, matching clinical TMB analysis pipelines.
Optimized for Illumina-based platforms (e.g., HiSeq X-TEN, as used in validation) and compatible with other NGS systems that support exome or targeted panel sequencing of the 39 Mb coding region. No additional protocol modifications are required for integration into existing workflows.
Used by kit manufacturers to validate the performance of new tTMB detection kits. During development, the standard can be spiked into normal gDNA to test if the kit maintains accuracy across different tumor content levels (e.g., confirming the kit can correctly measure TMB in 5%T samples without false lows).
Aids in setting kit specifications, such as defining the minimum tumor content required for reliable results (e.g., using the 2%T ratio to determine if the kit’s LOD meets clinical needs).
Clinical labs can use the standard to assess the stability of in-house TMB assays. For instance, testing the same batch of tTMB-P9 across multiple runs to verify inter-run reproducibility of TMB values at each tumor content ratio.
Supports comparison of different assay versions (e.g., updated probe panels or bioinformatics pipelines) by using the standard as a consistent reference to measure improvements in sensitivity or accuracy.
Serves as a positive control in daily TMB testing. Labs can run the standard alongside patient samples to monitor if the NGS workflow (e.g., DNA extraction, library preparation, sequencing) is functioning properly—if the standard’s TMB value deviates from the reference range, it indicates potential errors in the workflow.
Facilitates compliance with regulatory requirements: the standard provides traceable QC data, which is essential for meeting clinical laboratory accreditation standards (e.g., CAP, CLIA) for TMB testing.
The tTMB-P1 to tTMB-P8 standards are single-tumor-content (100%T) references with fixed TMB values (ranging from 5.37 to 27.15 mutations/Mb), designed primarily for calibrating TMB value ranges and verifying assay linearity. In contrast, tTMB-P9 is a matrix standard focused on tumor content variability (1%–10%T), targeting the evaluation of how low tumor cellularity impacts TMB measurement. The two types complement each other: P1-P8 ensure accurate TMB value assignment, while P9 ensures assay robustness in heterogeneous clinical samples.
No. The tTMB-P9 Standard is formulated with genomic DNA from tissue samples (gDNA) and is specifically designed for tissue TMB (tTMB) assay QC. For blood TMB (bTMB) testing, which uses circulating tumor DNA (ctDNA), Cobio Biotech offers dedicated bTMB standards (e.g., bTMB-P1), which are optimized for ctDNA’s unique characteristics (e.g., low concentration, fragmentation). Using tTMB-P9 for bTMB QC will not reflect the performance of ctDNA-specific workflows.
When stored at -20°C (avoiding repeated freeze-thaw cycles), the standard maintains stability for 12 months from the date of receipt. For short-term use (up to 1 month), it can be stored at 4°C. It is recommended to aliquot the standard into small volumes upon receipt to prevent degradation from repeated freeze-thaws.
The 1%T + 99%N ratio is the most challenging for TMB assays, as it contains extremely low levels of tumor-derived mutations. A reliable assay should detect a TMB value close to the reference (0.95 mutations/Mb at 2% AF cutoff). If the measured TMB is significantly lower than the reference, it indicates the assay may lack sufficient sensitivity for low-tumor-content samples. If the value is higher, it may suggest false-positive variant calls (e.g., from normal cell mutations or sequencing errors), requiring optimization of bioinformatics filters.
Every batch of tTMB-P9 Standard undergoes duplicate WES validation to confirm TMB value consistency (variation ≤ 5% from the reference range). Additionally, we test for DNA integrity (using agarose gel electrophoresis) and purity (A260/A280 ratio 1.8–2.0) to ensure the standard meets high-quality gDNA requirements—critical for reliable NGS performance.
The 1%–10% tumor content gradient is based on analysis of thousands of clinical tissue samples, addressing the most common issue labs face: inaccurate TMB results from low-tumor-content biopsies. By using this standard, labs can proactively identify and resolve workflow limitations before they affect patient test results.
We provide a detailed technical datasheet with reference TMB values for all tumor content ratios and AF cutoffs, as well as a recommended workflow for using the standard in QC. Our team of bioinformatics and molecular biology experts is also available to assist with data interpretation, assay optimization, and troubleshooting—ensuring labs get the most value from the product.
The tTMB-P9 Standard is developed in line with global regulatory guidelines for in vitro diagnostic (IVD) QC standards (e.g., FDA, NMPA). It has been used to support the development of NMPA-approved TMB detection kits (e.g., non-small cell lung cancer TMB kits), demonstrating its suitability for regulatory-compliant clinical workflows.
Name | tTMB-P9(matrix) Reference Standard |
Cat. No. | CBP80001-9 |
Format | Genomic DNA |
Size | 1ug+1ug |
Inventory Status | In Stock |
Buffer | Tris-EDTA |
Storage Conditions | 2~8℃ |
Expiry | 36 months from the date of manufacture |

Sample ID | Description | Cut off=1% | Cut off=2% | Cut off=3% | Cut off=4% | Cut off=5% |
DC208D0421 | 10%T+90%N | 22.94 | 19.72 | 16.53 | 12.50 | 8.41 |
DC208D0422 | 5%T+95%N | 15.75 | 10.50 | 5.82 | 2.92 | 1.37 |
DC208D0423 | 2%T+98%N | 8.29 | 2.12 | 0.75 | 0.42 | 0.21 |
DC208D0424 | 1%T+99%N | 4.15 | 0.95 | 0.27 | 0.18 | 0.09 |
500xWES; IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M; HiSeq X-TEN,af choose 0.01 cut off,call mutation,Filter again, and then calculate TMB according to the gradient cut off. Tumor sample: stage 4, adenocarcinoma Lung, Female Normal sample: B lymphoblast from the same individual | ||||||
Cat.No. | ID | TMB Value | Method |
CBP80001-1 | tTMB-P1 | 5.37 | WES |
CBP80001-2 | tTMB-P2 | 9.84 | WES |
CBP80001-3 | tTMB-P3 | 12.41 | WES |
CBP80001-4 | tTMB-P4 | 21.09 | WES |
CBP80001-5 | tTMB-P5 | 27.15 | WES |
CBP80001-6 | tTMB-P6 | 8.98 | WES |
CBP80001-7 | tTMB-P7 | 6.83 | WES |
CBP80001-8 | tTMB-P8 | 27.15 | WES |
CBP80001-10 | bTMB-P1 | 22.91 | WES |
General information
Name | tTMB-P9(matrix) Reference Standard |
Cat. No. | CBP80001-9 |
Format | Genomic DNA |
Size | 1ug+1ug |
Inventory Status | In Stock |
Buffer | Tris-EDTA |
Storage Conditions | 2~8℃ |
Expiry | 36 months from the date of manufacture |
General information
Name | tTMB-P9(matrix) Reference Standard |
Cat. No. | CBP80001-9 |
Format | Genomic DNA |
Size | 1ug+1ug |
Inventory Status | In Stock |
Buffer | Tris-EDTA |
Storage Conditions | 2~8℃ |
Expiry | 36 months from the date of manufacture |
Detection Methods
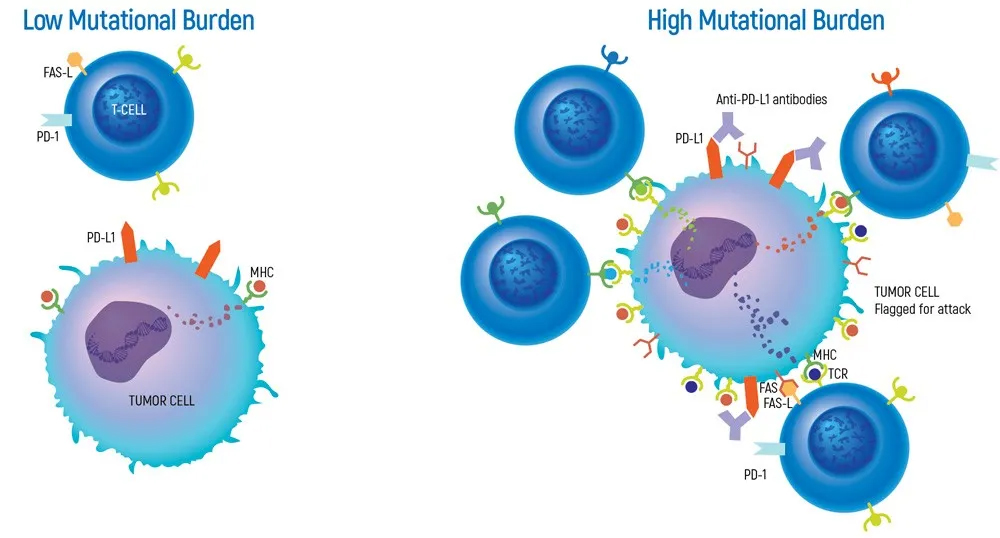
The current detection methods on the market refer to the number of somatic mutations per million bases (Mb) in the coding region of the patient's targeted sequencing, including point mutations and insertions and deletions. The number of somatic mutations in different cancers ranges from 0.01 mutations/Mb to more than 400 mutations/Mb.
The higher the tumor mutation load, the more likely the corresponding tumor-related carcinogenic mutations are, the more prominent the tumor's personality is, and the more different it is from normal cells.

Regarding TMB detection: Traditional detection technology is to analyze the patient's TMB by taking the patient's tumor tissue (tTMB). Currently, blood TMB (bTMB) can be detected, and compared with tissue detection, blood detection is more convenient and faster, and the non-invasive operation method also avoids more pain for patients.
Detection Methods
The current detection methods on the market refer to the number of somatic mutations per million bases (Mb) in the coding region of the patient's targeted sequencing, including point mutations and insertions and deletions. The number of somatic mutations in different cancers ranges from 0.01 mutations/Mb to more than 400 mutations/Mb.
The higher the tumor mutation load, the more likely the corresponding tumor-related carcinogenic mutations are, the more prominent the tumor's personality is, and the more different it is from normal cells.

Regarding TMB detection: Traditional detection technology is to analyze the patient's TMB by taking the patient's tumor tissue (tTMB). Currently, blood TMB (bTMB) can be detected, and compared with tissue detection, blood detection is more convenient and faster, and the non-invasive operation method also avoids more pain for patients.
Detailed Data
Sample ID | Description | Cut off=1% | Cut off=2% | Cut off=3% | Cut off=4% | Cut off=5% |
DC208D0421 | 10%T+90%N | 22.94 | 19.72 | 16.53 | 12.50 | 8.41 |
DC208D0422 | 5%T+95%N | 15.75 | 10.50 | 5.82 | 2.92 | 1.37 |
DC208D0423 | 2%T+98%N | 8.29 | 2.12 | 0.75 | 0.42 | 0.21 |
DC208D0424 | 1%T+99%N | 4.15 | 0.95 | 0.27 | 0.18 | 0.09 |
500xWES; IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M; HiSeq X-TEN,af choose 0.01 cut off,call mutation,Filter again, and then calculate TMB according to the gradient cut off. Tumor sample: stage 4, adenocarcinoma Lung, Female Normal sample: B lymphoblast from the same individual | ||||||
Detailed Data
Sample ID | Description | Cut off=1% | Cut off=2% | Cut off=3% | Cut off=4% | Cut off=5% |
DC208D0421 | 10%T+90%N | 22.94 | 19.72 | 16.53 | 12.50 | 8.41 |
DC208D0422 | 5%T+95%N | 15.75 | 10.50 | 5.82 | 2.92 | 1.37 |
DC208D0423 | 2%T+98%N | 8.29 | 2.12 | 0.75 | 0.42 | 0.21 |
DC208D0424 | 1%T+99%N | 4.15 | 0.95 | 0.27 | 0.18 | 0.09 |
500xWES; IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M; HiSeq X-TEN,af choose 0.01 cut off,call mutation,Filter again, and then calculate TMB according to the gradient cut off. Tumor sample: stage 4, adenocarcinoma Lung, Female Normal sample: B lymphoblast from the same individual | ||||||
Product List
Cat.No. | ID | TMB Value | Method |
CBP80001-1 | tTMB-P1 | 5.37 | WES |
CBP80001-2 | tTMB-P2 | 9.84 | WES |
CBP80001-3 | tTMB-P3 | 12.41 | WES |
CBP80001-4 | tTMB-P4 | 21.09 | WES |
CBP80001-5 | tTMB-P5 | 27.15 | WES |
CBP80001-6 | tTMB-P6 | 8.98 | WES |
CBP80001-7 | tTMB-P7 | 6.83 | WES |
CBP80001-8 | tTMB-P8 | 27.15 | WES |
CBP80001-10 | bTMB-P1 | 22.91 | WES |
Product List
Cat.No. | ID | TMB Value | Method |
CBP80001-1 | tTMB-P1 | 5.37 | WES |
CBP80001-2 | tTMB-P2 | 9.84 | WES |
CBP80001-3 | tTMB-P3 | 12.41 | WES |
CBP80001-4 | tTMB-P4 | 21.09 | WES |
CBP80001-5 | tTMB-P5 | 27.15 | WES |
CBP80001-6 | tTMB-P6 | 8.98 | WES |
CBP80001-7 | tTMB-P7 | 6.83 | WES |
CBP80001-8 | tTMB-P8 | 27.15 | WES |
CBP80001-10 | bTMB-P1 | 22.91 | WES |