- Home
- Products
- Services
- Resources
- About Us
- News
- Contact

loading
CBP80001-10
CBP80001-10
| Availability: | |
|---|---|
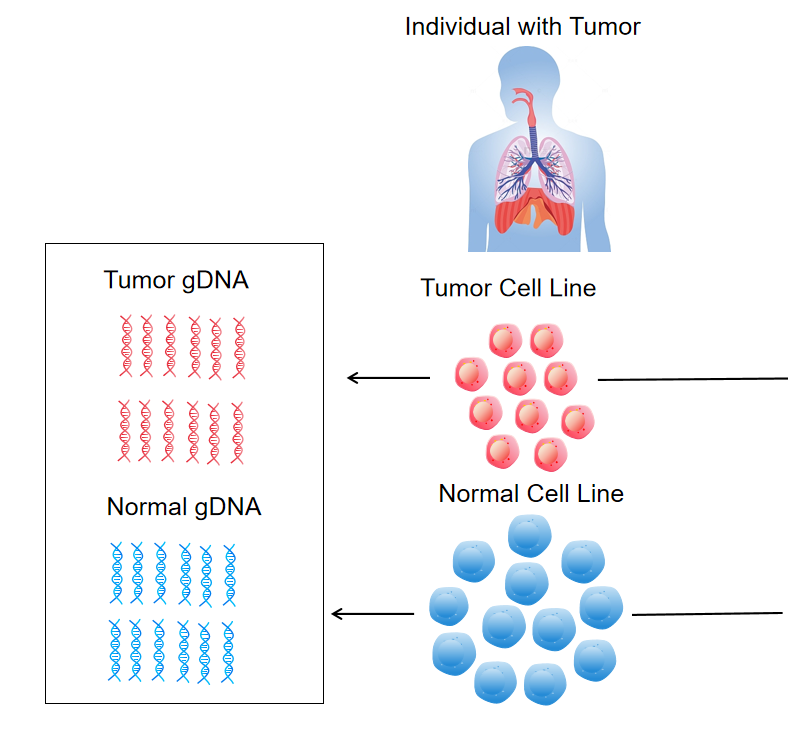
The Dual ctDNA/gDNA TMB 22.91 Standard for I-O NGS is a cutting-edge reference material engineered to validate tumor mutational burden (TMB) detection workflows in immuno-oncology (I-O) next-generation sequencing (NGS) applications. This reference standard combines genomic DNA (gDNA) with a precisely calibrated 22.91 mutations/Mb TMB value, representing the clinically relevant high-TMB phenotype associated with immune checkpoint inhibitor responsiveness. The cell-free tumor DNA (ctDNA) TMB 22.91 Reference Standard needs to be customized. Designed to mimic the complex mutational landscape of clinical specimens, this standard addresses the critical need for validated controls that span both liquid and tissue-based TMB assessment, enabling accurate assay calibration across the entire I-O diagnostic pipeline .
TMB Reference Standards are available in ctDNA (cell-free fraction) and gDNA (genomic fraction) formats to validate TMB workflows. The ctDNA component undergoes controlled fragmentation to 160-180 bp—mimicking the size distribution of circulating tumor DNA in plasma—while the gDNA maintains high molecular weight (>20kb) for tissue assay validation .
The 22.91 mutations/Mb value is verified by orthogonal methods including whole-exome sequencing (>500x coverage) and targeted panel sequencing, ensuring accuracy within ±15% of the nominal value. This level of precision enables reliable assessment of assay sensitivity for high-TMB detection, a critical factor in patient selection for immunotherapy .
The standard contains a comprehensive mutational profile including substitutions, indels, and copy number variations representative of high-TMB tumors. It covers >1,500 cancer-related genes with mutation types distributed to reflect real-world immuno-oncology cohorts, including actionable mutations in TP53, KRAS, and PD-L1 associated pathways .
For ctDNA workflow validation, dilute the standard to 10-20 ng/μL in plasma-derived cell-free DNA matrix. For gDNA applications, use at 20-50 ng/μL in normal genomic DNA background.
Incorporate the standard into validation runs to:
• Verify TMB calculation accuracy across targeted panels and WES
• Establish lower limit of detection for mutation calling
• Monitor inter-run variability in TMB scoring
• Validate bioinformatics pipelines for variant classification
Store unopened vials at -80°C for up to 24 months from manufacture date. After first use, aliquot remaining material into single-use volumes and store at -80°C to avoid freeze-thaw cycles. Thaw on ice for 15 minutes before use and mix gently by pipetting .
The 22.91 mutations/Mb TMB value falls within the clinically established high-TMB range associated with response to PD-1/PD-L1 inhibitors. Its dual matrix design allows validation of both tissue and liquid biopsy approaches commonly used in immuno-oncology research and clinical testing .
TMB is calculated as the total number of non-synonymous somatic mutations per megabase of sequenced DNA, excluding driver mutations and germline variants. This methodology aligns with FDA-recommended approaches for TMB assessment in clinical trials .
Yes, the standard is optimized for use with common oncology panels (50-500 genes). It includes a panel-specific TMB conversion factor in the certificate of analysis to enable accurate comparison between different panel sizes and designs .
No, the standard is depleted of common germline polymorphisms (MAF >1%) to ensure accurate distinction between somatic mutations and germline variants, a critical requirement for TMB calculation .
Name | bTMB-P1 (22.91) Reference Standard |
Cat. No. | CBP80001-10 |
Format | Genomic DNA |
Size | 1ug+1ug |
Inventory Status | In Stock |
Buffer | Tris-EDTA |
Storage Conditions | 2~8℃ |
Expiry | 36 months from the date of manufacture |
Detection Methods
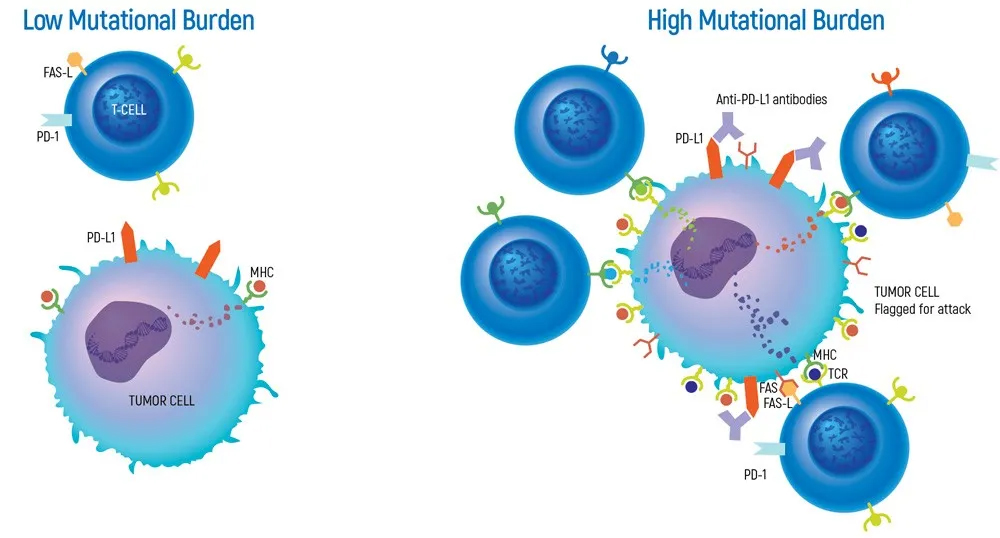
The current detection methods on the market refer to the number of somatic mutations per million bases (Mb) in the coding region of the patient's targeted sequencing, including point mutations and insertions and deletions. The number of somatic mutations in different cancers ranges from 0.01 mutations/Mb to more than 400 mutations/Mb.
The higher the tumor mutation load, the more likely the corresponding tumor-related carcinogenic mutations are, the more prominent the tumor's personality is, and the more different it is from normal cells.
Sample ID | Cat.No. | Format | Background | Assay | Comments |
DC20030079 | CBP80001-10N | gDNA | B lymphoblast, Female | 500x WES | Same individual |
DC20030078 | CBP80001-10T | ctDNA (Enzymatic digestion) | stage 4, adenocarcinoma Lung, Female | ||
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; DC20030078 is 100% Tumor sample and DC20030079 is 100% Normal sample. | |||||
Cut off =1% | Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 769 | 768 | 734 | 720 | 713 |
TMB value | 22.91 | 22.91 | 21.90 | 21.48 | 21.27 |
Cat.No. | ID | TMB Value | Method |
CBP80001-1 | tTMB-P1 | 5.37 | WES |
CBP80001-2 | tTMB-P2 | 9.84 | WES |
CBP80001-3 | tTMB-P3 | 12.41 | WES |
CBP80001-4 | tTMB-P4 | 21.09 | WES |
CBP80001-5 | tTMB-P5 | 27.15 | WES |
CBP80001-6 | tTMB-P6 | 8.98 | WES |
CBP80001-7 | tTMB-P7 | 6.83 | WES |
CBP80001-8 | tTMB-P8 | 27.15 | WES |
CBP80001-9 | tTMB-P9 | N/A | WES |
The Dual ctDNA/gDNA TMB 22.91 Standard for I-O NGS is a cutting-edge reference material engineered to validate tumor mutational burden (TMB) detection workflows in immuno-oncology (I-O) next-generation sequencing (NGS) applications. This reference standard combines genomic DNA (gDNA) with a precisely calibrated 22.91 mutations/Mb TMB value, representing the clinically relevant high-TMB phenotype associated with immune checkpoint inhibitor responsiveness. The cell-free tumor DNA (ctDNA) TMB 22.91 Reference Standard needs to be customized. Designed to mimic the complex mutational landscape of clinical specimens, this standard addresses the critical need for validated controls that span both liquid and tissue-based TMB assessment, enabling accurate assay calibration across the entire I-O diagnostic pipeline .
TMB Reference Standards are available in ctDNA (cell-free fraction) and gDNA (genomic fraction) formats to validate TMB workflows. The ctDNA component undergoes controlled fragmentation to 160-180 bp—mimicking the size distribution of circulating tumor DNA in plasma—while the gDNA maintains high molecular weight (>20kb) for tissue assay validation .
The 22.91 mutations/Mb value is verified by orthogonal methods including whole-exome sequencing (>500x coverage) and targeted panel sequencing, ensuring accuracy within ±15% of the nominal value. This level of precision enables reliable assessment of assay sensitivity for high-TMB detection, a critical factor in patient selection for immunotherapy .
The standard contains a comprehensive mutational profile including substitutions, indels, and copy number variations representative of high-TMB tumors. It covers >1,500 cancer-related genes with mutation types distributed to reflect real-world immuno-oncology cohorts, including actionable mutations in TP53, KRAS, and PD-L1 associated pathways .
For ctDNA workflow validation, dilute the standard to 10-20 ng/μL in plasma-derived cell-free DNA matrix. For gDNA applications, use at 20-50 ng/μL in normal genomic DNA background.
Incorporate the standard into validation runs to:
• Verify TMB calculation accuracy across targeted panels and WES
• Establish lower limit of detection for mutation calling
• Monitor inter-run variability in TMB scoring
• Validate bioinformatics pipelines for variant classification
Store unopened vials at -80°C for up to 24 months from manufacture date. After first use, aliquot remaining material into single-use volumes and store at -80°C to avoid freeze-thaw cycles. Thaw on ice for 15 minutes before use and mix gently by pipetting .
The 22.91 mutations/Mb TMB value falls within the clinically established high-TMB range associated with response to PD-1/PD-L1 inhibitors. Its dual matrix design allows validation of both tissue and liquid biopsy approaches commonly used in immuno-oncology research and clinical testing .
TMB is calculated as the total number of non-synonymous somatic mutations per megabase of sequenced DNA, excluding driver mutations and germline variants. This methodology aligns with FDA-recommended approaches for TMB assessment in clinical trials .
Yes, the standard is optimized for use with common oncology panels (50-500 genes). It includes a panel-specific TMB conversion factor in the certificate of analysis to enable accurate comparison between different panel sizes and designs .
No, the standard is depleted of common germline polymorphisms (MAF >1%) to ensure accurate distinction between somatic mutations and germline variants, a critical requirement for TMB calculation .

Name | bTMB-P1 (22.91) Reference Standard |
Cat. No. | CBP80001-10 |
Format | Genomic DNA |
Size | 1ug+1ug |
Inventory Status | In Stock |
Buffer | Tris-EDTA |
Storage Conditions | 2~8℃ |
Expiry | 36 months from the date of manufacture |
Detection Methods
The current detection methods on the market refer to the number of somatic mutations per million bases (Mb) in the coding region of the patient's targeted sequencing, including point mutations and insertions and deletions. The number of somatic mutations in different cancers ranges from 0.01 mutations/Mb to more than 400 mutations/Mb.
The higher the tumor mutation load, the more likely the corresponding tumor-related carcinogenic mutations are, the more prominent the tumor's personality is, and the more different it is from normal cells.

Sample ID | Cat.No. | Format | Background | Assay | Comments |
DC20030079 | CBP80001-10N | gDNA | B lymphoblast, Female | 500x WES | Same individual |
DC20030078 | CBP80001-10T | ctDNA (Enzymatic digestion) | stage 4, adenocarcinoma Lung, Female | ||
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; DC20030078 is 100% Tumor sample and DC20030079 is 100% Normal sample. | |||||
Cut off =1% | Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 769 | 768 | 734 | 720 | 713 |
TMB value | 22.91 | 22.91 | 21.90 | 21.48 | 21.27 |
Cat.No. | ID | TMB Value | Method |
CBP80001-1 | tTMB-P1 | 5.37 | WES |
CBP80001-2 | tTMB-P2 | 9.84 | WES |
CBP80001-3 | tTMB-P3 | 12.41 | WES |
CBP80001-4 | tTMB-P4 | 21.09 | WES |
CBP80001-5 | tTMB-P5 | 27.15 | WES |
CBP80001-6 | tTMB-P6 | 8.98 | WES |
CBP80001-7 | tTMB-P7 | 6.83 | WES |
CBP80001-8 | tTMB-P8 | 27.15 | WES |
CBP80001-9 | tTMB-P9 | N/A | WES |
General information
Name | bTMB-P1 (22.91) Reference Standard |
Cat. No. | CBP80001-10 |
Format | Genomic DNA |
Size | 1ug+1ug |
Inventory Status | In Stock |
Buffer | Tris-EDTA |
Storage Conditions | 2~8℃ |
Expiry | 36 months from the date of manufacture |
General information
Name | bTMB-P1 (22.91) Reference Standard |
Cat. No. | CBP80001-10 |
Format | Genomic DNA |
Size | 1ug+1ug |
Inventory Status | In Stock |
Buffer | Tris-EDTA |
Storage Conditions | 2~8℃ |
Expiry | 36 months from the date of manufacture |
Detection Methods
The current detection methods on the market refer to the number of somatic mutations per million bases (Mb) in the coding region of the patient's targeted sequencing, including point mutations and insertions and deletions. The number of somatic mutations in different cancers ranges from 0.01 mutations/Mb to more than 400 mutations/Mb.
The higher the tumor mutation load, the more likely the corresponding tumor-related carcinogenic mutations are, the more prominent the tumor's personality is, and the more different it is from normal cells.

Regarding TMB detection: Traditional detection technology is to analyze the patient's TMB by taking the patient's tumor tissue (tTMB). Currently, blood TMB (bTMB) can be detected, and compared with tissue detection, blood detection is more convenient and faster, and the non-invasive operation method also avoids more pain for patients.
Detection Methods
The current detection methods on the market refer to the number of somatic mutations per million bases (Mb) in the coding region of the patient's targeted sequencing, including point mutations and insertions and deletions. The number of somatic mutations in different cancers ranges from 0.01 mutations/Mb to more than 400 mutations/Mb.
The higher the tumor mutation load, the more likely the corresponding tumor-related carcinogenic mutations are, the more prominent the tumor's personality is, and the more different it is from normal cells.

Regarding TMB detection: Traditional detection technology is to analyze the patient's TMB by taking the patient's tumor tissue (tTMB). Currently, blood TMB (bTMB) can be detected, and compared with tissue detection, blood detection is more convenient and faster, and the non-invasive operation method also avoids more pain for patients.
Detailed Data
Sample ID | Cat.No. | Format | Background | Assay | Comments |
DC20030079 | CBP80001-10N | gDNA | B lymphoblast, Female | 500x WES | Same individual |
DC20030078 | CBP80001-10T | ctDNA (Enzymatic digestion) | stage 4, adenocarcinoma Lung, Female | ||
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; DC20030078 is 100% Tumor sample and DC20030079 is 100% Normal sample. | |||||
Cut off =1% | Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 769 | 768 | 734 | 720 | 713 |
TMB value | 22.91 | 22.91 | 21.90 | 21.48 | 21.27 |
Detailed Data
Sample ID | Cat.No. | Format | Background | Assay | Comments |
DC20030079 | CBP80001-10N | gDNA | B lymphoblast, Female | 500x WES | Same individual |
DC20030078 | CBP80001-10T | ctDNA (Enzymatic digestion) | stage 4, adenocarcinoma Lung, Female | ||
IDT xGenExome Research Panel v1.0capture,Target Region 39M, Probe Region 51M,CDS Region 33.52M; DC20030078 is 100% Tumor sample and DC20030079 is 100% Normal sample. | |||||
Cut off =1% | Cut off =2% | Cut off =3% | Cut off =4% | Cut off =5% | |
Mutation Number | 769 | 768 | 734 | 720 | 713 |
TMB value | 22.91 | 22.91 | 21.90 | 21.48 | 21.27 |
Product list
Cat.No. | ID | TMB Value | Method |
CBP80001-1 | tTMB-P1 | 5.37 | WES |
CBP80001-2 | tTMB-P2 | 9.84 | WES |
CBP80001-3 | tTMB-P3 | 12.41 | WES |
CBP80001-4 | tTMB-P4 | 21.09 | WES |
CBP80001-5 | tTMB-P5 | 27.15 | WES |
CBP80001-6 | tTMB-P6 | 8.98 | WES |
CBP80001-7 | tTMB-P7 | 6.83 | WES |
CBP80001-8 | tTMB-P8 | 27.15 | WES |
CBP80001-9 | tTMB-P9 | N/A | WES |
Product list
Cat.No. | ID | TMB Value | Method |
CBP80001-1 | tTMB-P1 | 5.37 | WES |
CBP80001-2 | tTMB-P2 | 9.84 | WES |
CBP80001-3 | tTMB-P3 | 12.41 | WES |
CBP80001-4 | tTMB-P4 | 21.09 | WES |
CBP80001-5 | tTMB-P5 | 27.15 | WES |
CBP80001-6 | tTMB-P6 | 8.98 | WES |
CBP80001-7 | tTMB-P7 | 6.83 | WES |
CBP80001-8 | tTMB-P8 | 27.15 | WES |
CBP80001-9 | tTMB-P9 | N/A | WES |